ABSTRACT
TO acquire the phage-displayed mimotopes which mimic the specificity of hepatitis B virus surface antigen (HBsAg), a random peptide library expressing linear peptide with 12 amino acids in length were used to screen with the serum from a hepatitis B virus infected patient in the recovery phase. After 3 rounds of biopanning, the positive phages were confirmed by competitive ELISA using HBsAg/P33. Two phagotopes were identified and one of them was confirmed as mimotope by competition experiment. Based on the mimotpe, a multiple antigenic peptide with four branches was synthesized by solid phase peptide synthesis. The antiginicity and specificity of the synthesized antigen was tested in BALB/c mice compared with the native epitope-based antigen. The results showed that the mimotope-based antigen could evoke higher titer of antibodies with the same specificity of the epitope-based antigen. Those findings indicate mimotopes can be used in antigen and vaccine design.
Similar content being viewed by others
INTRODUCTION
Hepatitis B virus (HBV) infection is the most common viral infection in humans especially in China and East Asia. At present, the most efficient method to control the disease is vaccination of new-borns. Both blood derived vaccine and recombinant vaccine are hepatitis B surface antigen (HBsAg)-based and useful in the prevention of the disease1, 2, 3. HBsAg could evoke protective humoral immune response in vivo, but the immune memory only last for 5 years or so3, 4. How to design a more efficient vaccine to improve the immunity is still a problem. The HBsAg contain three polypeptides termed S (small), M (middle), and L (large), which result from the variable use of three start codons within the open reading frame of the surface antigen gene. The M polypeptide comprises the Pre-S2 sequence as its N-terminal region, followed by the S sequence. Epitopes in the Pre-S2 domain have been identified using a panel of monocolonal antibodies and peptides5, 6.
The hypothesis of our work is that an ideal protective immune response is based on a set of epitopes. Different combination of epitopes could induce different characteristics of immune response. To build an immunogen by using epitopes we established a new strategy, which is called epitope-based vaccine design (EBVD)7. In light of EBVD, we designed more than 200 vaccine candidates of hepatitis B. One of them was proved to be very powerful in termination of persistant infection of HBV, and another three were very efficient in evoking high titers of neutralizing antibodies8, 9. Those vaccine candidates are designed using natural epitopes identified by epitope mapping and mimotopes identified by phage-display and combinational chemistry. In this paper we report a mimotope identified from a phage-displayed library through the serum of a patient with viral hepatitis B.
MATERIALS AND METHODS
Reagents
Bacteria stain XL1-Blue [FproAB, lacIqz M15, Tn10(tetr)] and phagemid M13LP67 were provided by Dr.Devlin J J, Cetus Corp. The helper phage VSAM 13 and the murine monoclonoal antibody against M13 were purchased from Promega. Polyclonal sheep anti-human IgG biotinated antibodies and polyclonal sheep anti-mouce IgG peroxidase-conjugated antibodies were purchased from Biodin Corp (Beijing, China).
Construction of phage-displayed peptide library
Two complementary oligo nucleotides that encode all possible forms of linear peptides with the sequences 5′CTTTCTATTCTCT(NNB) 12 GGTGGAGGTTC3′ (P1) and 5′GGCCGAACCTCCACC(VNN)12AGAGTGAGAATAGAAAGGTAC-3′ (P2) were synthesized, in which N is A, T, C or G; B is G, T or C; V is G, C or A. To anneal the complementary oligonucleotides, 7 μg of P1 and 8 μg of P2 were phosphorylated with T4 kinase and mixed in 100 μl of 1 M NaCl. The mixture was heated to 95°C for 5 min and cooled slowly to 15°C. The phaghemid M13LP67 was digested with KpnI and EagI (45 μg/μl ). 20 μg of the completely digested phagemid were eluted from the agrose gel and mixed with 10-fold molar excess of annealed oligonucleotides. These fragments were ligated by addition of T4 ligase. The ligated DNA was introduced into Escherichia coli XL1-Blue by electroporation using the Gene Pulser (BioRad), and the transformed cells were infected with VSAM13 helper phage to amplify the peptide-displayed phage library.
Screening of mimotopes from a phage-displayed peptide library
To select the phages that display the candidate HBsAg mimotopes, the following biopanning protocol was adopted. Both HBsAb negtive normal human serum for nonspecific elution and a HBsAb positive patient's serum for specific elution of the phage were used for biopanning. A patient of 28-yr-old (HLA-A2, B44, and Cw5.Cw7) with acute viral hepatitis B was in recovery phase and the HBV marker in the serum is only HBsAb positive. The helper phage VSAM13 (1012pfu) inactivated by ultraviolet irradiation was incubated overnight with 10 μl normal human serum and then 10 μl of a phage-displayed peptide liberary (0.9×108pfu) was added and incubated for 8 h. An ELISA plate (96-well; Maxisorb, Nnuc) was coated with streptoavidin and incubated with polyclonal sheep anti-human IgG biotinated antibodies. The pre-treated helper phage and phage displayed library reacted with antibodies in normal human serum were captured. The supernatant containing phages not reacting with normal serum was harvested and captured by patient's serum. The patient's serum captured phages were competively eluted by biotin. The phages in all fractions were titered to determine the degree of selection during panning.
Phage ELISA for identification of positive phages
An ELISA plate was coated with properly diluted patient's HBsAb positive serum or nomal HBsAb negtive serum by incubation at 4°C overnight and then blocked with 5% BSA in PBS. Affinity selected phages were added to the wells and allowed to bind at 37°C for 1 hr. After the unbouned phages were removed with PBST, the bound phages were detected by incubation with murine anti-M13 antibodies (Promega) followed by anti-murine antibodies cogugated with horseradish peroxidase. The bound horseradish peroxidase was determined by incubation with 3 mg/ml of O-phenylenediamine dihydrochloride (Pierce Chemicals) in buffer (30 mM citrate, 70 mM Na2HPO4, and 0.02% H2O2, pH 5.5). After the reaction was stoped by the addition of 3N HCl, A492 nm was determined in an ELISA reader (model 550, Bio Rad). All the assays were carried out in triplicate. If Ap-An > 0.2, then it is considered as positive phages (Ap represents A492 for patient's serum, An represents A492 for the parallel normal serum).
To confirm the positive phage-displayed peptides are Pre-S2 region of HBsAg specific, a competitive ELISA employing recombinant HBsAg (HBsAg/P33, ayw subtype, kindly provided by Dr. Toillais P, Institut Pasteur) as the phage competitor was conducted.
DNA sequencing
The phages selected from the third round of biopanning were precipitated with polyethylene glycol after amplification and single-strand DNA was prepared by phenol extraction. The nucleotide sequence of the phagotope of the selected phages was determined by the dideoxynucleotide chain termination method(10) using the primer with the sequence 5′ -CCCTCATAGTTAGCGTAACG- 3′.
Peptide synthesis
A linear peptide with the single code sequence TANGFYRLPSGS and its octamer multiple- antigen peptides with the lysine core (MAP1, Fig 1) were synthesized respectively using solid- phase peptide synthesis method on the 431A model of peptide synthesizer (Perkin-Elmer). Control octamer multiple-antigen peptides based on the epitope of 130-150 of HBsAg (MAP2, Fig 1) was synthesized by same protocol. The peptides were purified by high performance liquid chromatography (HPLC) and lyophilized. The purity of the peptides was 98.5% or more assessed by C18 reverse-phase high-pressure liquid chromatography.
Animals and immunization
Inbred female BALB/c (H-2d) mice were purchased from the Animal Center of the University, Chongqing, P. R. China.
Groups of 6- to 7-week-old BALB/c mice were immunized i.p. with 5 nM of MAP1 or MAP2 in Freund's complete adjuvant (CFA). The mice were boosted 3 weeks later with the same dose of peptides in Freund's incomplete adjuvant, and sera were collected weekly. Further groups of mice received a second boost (5 nM in Freund's incomplete adjuvant) 6 w after the first boost.
Immunochemical assays
Peptide ELISA. 96-well microplates (Nunc, Roskilde, Denmark) were coated with 50 μl of peptides at 5 μg /ml in sodium bicarbonate buffer (pH 9.6) overnight at 4°C. The plates were washed with water, and blocked with 200 ml of phosphate-buffered saline (PBS)-bovine serum albumin (BSA) 2.5% per well for 2 h at 37°C. After being washed, the plates were incubated with twofold serial dilutions of biotinylated anti-HbsAg/p33 Mab or polyclonal murine anti-peptide serum in PBS-Tween (PBS-T)-BSA 2.5 % (diluting buffer) for 2 h at 37°C and washed.
For biotinylated Mab, plates were incubated for 1 h at 37°C with a steptavidin-peroxidase conjugate (Boehringer, Mannheim, Germany) at 1/5,000 in diluting buffer. For polyclonal sera, plates were incubated with a rabbit anti-mouse IgG (heavy and light chains)-peroxidase conjugate (Nordic, Tillburg, The Netherlands) at 1/2,000 in diluting buffer for 1 hr at 37°C.
Unbound conjugate was removed by washing, and enzyme activity was detected by the addition of 100 μl of substrate solution (o-phenylenediamine-0.004% hydogen peroxide in citrate- phosphate buffer). After 10 min at room tempreture the enzymatic reaction was stoped with 50 μl of 2 M H2SO4. IgG titers were expressed as log10 of the reciprocal of the dilution giving an absorbence at 492 nm more than 0.2.
Reactivity of Mab25-19 (kindly provided by Dr. Mimms) with the linear peptides, MAP1 and MAP2, was assessed by a Mab25-19 based ELISA and a competitive ELISA based on peptide binding with Mab25-19 as the competitor of immunized mouse sera.
RESULTS
A phage-displayed peptide library composed of 2.1×108independent recombinants was constructed. Each phage displays a 12 amino acid-lengh of peptide as a fusion protein with the NH2 terminus of gIII coat protein.
Selection of the phages that display mimotopes of HBsAg from a phage-displayed peptide library
A combination of nomal human sera for nonspecific elution and HBsAg for the specific elution of the phages bound to patient's serum was used to select the phages that display the potential mimotopes. The result of three rounds of biopanning by patient's serum is shown in 20 randomly chosen phage clones that were eluted with 5 μM of HBsAg during the third round of biopanning were amplified and purified for further study.
Binding of affinity-selected phages to patient's serum
In a sandwich ELISA with patient's serum, two clones, A and B, were found to bind with patient's serum while B has higher absorbence. (Fig 2) phages were furtherly screened by HBsAg competition ELISA. HBsAg significantly inhibited the binding of Clone B to patient's serum while the binding of clone A was not significantly influenced by HBsAg (lower).
Clone A was not considered as mimotope bearing phage but clone B was. By using the method mentioned in Materials and methods, the nucleotide determined as ACTGCTAATGGTTTTTATAGGCTGCCTTCTGGTTCG and the amino acid sequence is deduced as TANGFYRLPSGS. Analysis of homology of the phagotope to HBsAg was conducted using ANTHEPROT V2.9. The amino acid sequence of the peptide is similar to 135-146 of Pre-S2 region of HBsAg which had been proved as a protective epitope and recognized by Mab25-1911, 12.
Induction of specific anti-HBs antibody response
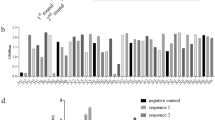
To assess the immunogenicity of the HBsAg mimotpe presented as MAP constructs, BALB/c mice were immunized with the constructs, and peak anti-peptide antibody titers (log10 tier range) were observed 4 to 5 w after the boost. Sera obtained from mice immunized with either MAP1 or MAP2 construct at the peak of antibody response were tested for their abilities to cross- react with HBsAg/P33 and to compete with Mab 25–19. Although both MAP1 and MAP2 induced antibodies cross-react with HBsAg/P33, MAP1 generated higher titer (log10 titer, 3.18±4.18) (Fig 4). Furthermore, MAP1 induced the higher titer of antibodies that specifically inhibited the binding of Mab25-19 to HBsAg (Fig 5).
Quantitative inhibition of HBsAg-specific Mab 25-19 binding by anti-mimotpe (MAP1) and anti-epitope antisera (MAP2) from immunized Balb/c mice. The antisera were collected at fifth week after boost. The positive cut off value was A492≥0.2. The negative control wells were using sera from unimmunized mice.
DISCUSSION
Phage displayed library was primarily used to murine monoclonal antibody-based biopanning of mimotopes. But in recent years, several mimotopes were identified through polyclonal antisera screening of phage displayed library13, 14, 22. It was reported that a mimotope related to S protein of HBsAg by using 2 different sera from immunized individuals and phage-displayed random peptide liberary15. In this paper we report a new mimotope which is related with Pre-S2 region of M protein of HBsAg by using one patient's serum strategy.
We constructed a 12 amino acids phage peptide library with 2.1×108 independent recombinants. Although this represents a small portion of possible diversity (416×212=1.75×1013), the diversity of our library was still large enough for screening. After three rounds of biopanning, one clone of phage was identified as mimotope bearing phage. Several lines of experimental evidence presented in this report suggest that the selected type A phages specifically bind to anti-Pre-S2 antibodies through its NH2-terminal epitope peptide. A new mimotope of HBsAg was identified by polycolonal human sera. This indicates that using patient's sera can screen mimotopes. A major goal of the entire strategy was to increase the efficiency of selection of disease-specific clones and to identify them without using the antigen. In reference 15, the positive sera were from vaccine immunized individuals and more than one patient serum were used in the immunosceening step. Thus the mimotope identified by that strategy might be more closely related with antibodies for prevention but less related with antibodies for disease recovery. In this report, we used a patient serum in early recovery phase in the immunoscreening step. Therefore, the screened mimotope may be closely related with disease recovery. In the one patient serum strategy, the screening step using large panels of negative sera is more immportant, and the mimotope must be verified by Mabs with clearly epitope background.
The branched form of phage A mimotope peptide was synthesized according to the DNA sequence of the NH2-terminal region of the gIII gene of the phage. There are six amino acids in the mimotope same to that in the native sequence in Pre-S2 region of HBsAg. In a former study, we found that R137 and A143 were critical for the binding to Mab25-19 by pepscan12. Whoever, this study indicates N and S could replace R137 and A143 respectively. This finding implied that the new combination of amino acids in the mimotope may consitute a new epitope with the same specificity. Another possibility is the mimotope identified in this report is just a cross reactive structure of the native epitope in the Pre-S2 region of HBsAg.
It is definitely known that Mab25-19 aginst the native epitope in the Pre-S2 region of HBsAg is a neutralizing and protective antibody16, 17. It rises at the early stage of hepatitis B virus infection in humans and closely related with prognosis of the disease18. It has been recommended by many authors that this epitope should be brought to components of new generation of HB preventive vaccine16, 19, 20, 21. The results of competition tests employing Mab 25-19 indicated that the mimotope identified in this report was a protective epitope and could be considered in the HB vaccine design.
References
Lauderdale, DS, Oram RJ, Goldstein KP, Daum AS . Hepatitis B vaccination among children in inner-city public housing. JAMA 1999; 282:1725.
Bertolino JG . Newborn hepatitis B immunization rates in primary care practices. Arch Pedistr Adolesc Med 1996; 150:1173.
Dobson S, Scheifele D, Bell A . Assessment of a universal, school-based hepatitis B vaccination program. 1995; JAMA 274:1209.
Del Canho, R., Grosheide, PM, Geijtink, RA, Schalm SW . The course of hepatitis B in 9 children who become positive for viral surface antigen in spite of vaccination. Ned Tijdschr Geneeskd 1993; 137:2599.
Lee MK, Kim, KL, Hahm KS . Epitope mapping of Pre-S (2) of the hepatitis B virus surface antigen against a conformation-dependent monoclonal antibody using synthetic peptides. Biochem Mol Biol Int 1996; 40:1077.
Meisel H, Sominskaya I, Pumpens P, Pushko P, Borisova G, Deepen R, Lu X, Spiller GH, Kruger DH, Grens E . Fine mapping and functional characterization of two immuno- dominant regions from the sequence of hepatitis B virus. Intervirology 1994; 37:330.
Wu Y, Jia Z, Zou L, Shih T . EBVD-a new approach to vaccine design. The Immunologist 1998; 6(s):1487.
Shih T, Wu Y . Molecular design and immunological investigations on the therapeutic peptide vaccines against viral hepatitis B. The Immunologist 1998; 6(4):947.
Wu Y, Liu M, Jia Z, Zou L . Studies on molecular design, chemical synthesis and immunogenicity of novel antigenic peptide against hepatitis B virus. Acta Academiae Medicine MiltarisTertiae 2000; 22:919.
Sanger F, Nicklen S, Coulson A . DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 1977; 74:5463.
Wu Y, Zhu X, Zhang L . The humoral response to Pre-(2) region of the surface antigen of hepatitis b virus. Immunol J 1993; 9:8.
Wu Y, Zhu X, Zhang L . The fine specificity of antibody binding sites of Pre-S(2) region of the surface antigen of hepatitis B virus. Immunol J 1994; 10:80.
Fierabracci A, Biro PA, Yiangou Y et al. Osteopontin is an autoantigen of the somatostatin cells in human islets: identification by screening random peptide libraries with sera of patients with insulin-dependent diabetes mellitus. Vaccine 1999; 18:342.
Lea IA, Adoyo P, O'Rand MG . Autoimmunogenicity of the human sperm protein Sp17 in vasectomized men and identification of linear B cell epitopes. Fertil Steril 1997; 67:355.
Folgori A, Tafi R, Meola A et al. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J 1994; 13:2236.
Mimms LT, Floreani, M, Tyner J et al. Discrimination of hepatitis B virus (HBV) subtypes using monoclonal antibodies to the PreS1 and PreS2 domains of the viral envelope. Virology. 1990; 174:604.
Heijtink RA, de-Wilde GA, van-Hattum J et al. Long-term immune reactivity to pre-S (2)-antigen after acute hepatitis B infection. J Med Virol. 1989; 27:95.
Galan MI, Tomas J, Bernal MC et al. Evaluation of the pre-S (pre-S (1) Ag/pre-S (2) Ab) system in hepatitis B virus infection. J Clin Pathol. 1991 J; 44:25.
Milich DR, Jones JE, McLachlan A et al. Importance of subtype in the immune response to the pre-S (2) region of the hepatitis B surface antigen. II. Synthetic Pre-S (2) immunogen. J Immunol. 1990 May 1; 144(9):3544.
Langley KE, Egan KM, Barendt JM et al. Characterization of purified hepatitis B surface antigen containing pre-S (2) epitopes expressed in Saccharomyces cerevisiae. Gene 1988; 67:229.
Itoh Y, Takai E, Ohnuma H et al. A synthetic peptide vaccine involving the product of the pre-S (2) region of hepatitis B virus DNA: protective efficacy in chimpanzees. Proc Natl Acad Sci USA 1986; 83:9174.
Yu ZZ et al. PSMA mimotope isolated from phage display peptide library can induce PSMA specific immune response. Cell Res 1999; 9:271.
Acknowledgements
This work was supported in part by Grants 39789010 and 39700132 from the Natural Science Foundation of China and agrant from National Education Ministery under the State Concil of China.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhang, W., WAN, Y., LI, D. et al. A mimotope of Pre-S2 region of surface antigen of viral hepatitis B screened by phage display. Cell Res 11, 203–208 (2001). https://doi.org/10.1038/sj.cr.7290087
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290087
Keywords
This article is cited by
-
Phage display as a promising approach for vaccine development
Journal of Biomedical Science (2016)
-
Mimotopes selected with a neutralizing antibody against urease B from Helicobacter pyloriinduce enzyme inhibitory antibodies in mice upon vaccination
BMC Biotechnology (2010)