Abstract
Telomerase reactivation is a hallmark of human carcinogenesis. Increased telomerase activity may result from gene amplification and/or overexpression. This study evaluates the prognostic value of hTERT gene amplification and mRNA overexpression in 144 resectable non-small-cell lung cancer (NSCLC) specimens. The hTERT gene copy number was assessed by quantitative polymerase chain reaction (qPCR) on laser-capture microdissected tumour cells of 81 tumours, and by fluorescence in situ hybridisation (FISH) on a subset of 59 tumours. hTERT mRNA level was determined by reverse transcription (RT)–qPCR in 130 tumours. In total, 57% of (46 out of 81) primary NSCLC specimens demonstrated hTERT amplification, which was significantly more common (P<0.001) in adenocarcinoma (30 out of 40) than in squamous cell carcinoma (13 out of 37). The hTERT mRNA overexpression was noted in 74% (94 out of 130) of tumours; it was more frequent in squamous cell than in adenocarcinoma (87 vs 68%, P=0.03). Overexpression was significantly associated with amplification (P=0.03), especially in adenocarcinoma. The hTERT gene amplification was prognostic for shorter recurrence-free survival (hazard ratio=2.16, P=0.03). These data indicate that gene amplification is an important mechanism for hTERT overexpression in lung adenocarcinoma and is an independent poor prognostic marker for disease-free survival in NSCLC.
Similar content being viewed by others
Main
Telomerase is a ribonucleoprotein reverse transcriptase complex containing an RNA subunit hTERC and a protein catalytic subunit hTERT (Nakamura et al, 1997). The hTERC RNA is expressed universally in eukaryotic cells and hTERT expression has been correlated with activation of the telomerase complex (Nakamura et al, 1997; Kolquist et al, 1998; Arinaga et al, 2000). Telomerase activity is absent in most human adult somatic cells. With continuous cell proliferation, there is a progressive loss of telomeric DNA that ultimately may trigger replicative senescence. The forced expression of hTERT cDNA in normal human cells has led to extension of the replicative lifespan (Vaziri and Benchimol, 1998). An alternative mechanism for cellular immortalisation is by the telomerase-independent ALT pathway (Newbold, 2002). Activations of the hTERT or ALT pathways are obligate for senescence bypass and for neoplastic transformation of normal cells (Newbold, 2002). Telomerase activity and/or hTERT expression are increased in cancers and are prognostic factors in various cancer types (Harada et al, 1999; Bieche et al, 2000; Lee et al, 2001; Marchetti et al, 2002; Wang et al, 2002; Fujita et al, 2003; Krams et al, 2003; Ohali et al, 2003; Tchirkov et al, 2003; Lantuejoul et al, 2004). However, the clinical impact of hTERT expression or activity in non-small-cell lung cancer (NSCLC) remains controversial (Albanell et al, 1997; Taga et al, 1999; Hirashima et al, 2000; Komiya et al, 2000; Toomey et al, 2001; Marchetti et al, 2002; Wang et al, 2002; Fujita et al, 2003; Wu et al, 2003; Hsu et al, 2004; Lantuejoul et al, 2004). The clinical significance of increased hTERT gene copy number has not been investigated.
Multiple mechanisms may regulate hTERT expression and activity. There is considerable evidence that transcriptional activation plays a major role in regulating hTERT mRNA expression (Ducrest et al, 2002), and the latter is correlated with telomerase activity (Arinaga et al, 2000; Marchetti et al, 2002; Saretzki et al, 2002). However, post-translational modifications may also contribute to the regulation of hTERT activity (Kang et al, 1999). Increased expression of hTERT resulting from gene amplification was recently reported in embryonal brain tumours (Fan et al, 2003) and cervical carcinoma (Zhang et al, 2002). The hTERT gene is located on chromosome 5p15, a chromosomal arm that is commonly overepresented or amplified in lung cancer (Luk et al, 2001). Amplification of the hTERT gene and a concomitant increase in telomerase activity has been reported in lung cancer cell lines and other cancer types (Zhang et al, 2000; Saretzki et al, 2002). In this study, we have investigated the frequency and prognostic significance of hTERT gene amplification and overexpression in NSCLC.
Materials and methods
Patients and clinical samples
Patients included in this study had undergone lobectomy or pneumonectomy for resection of their primary lung cancer but had not received prior radiation or chemotherapy. Altogether, 169 tissue samples from 144 patients were used; these included 144 tumours and a corresponding subset of 30 non-neoplastic lung tissues. The latter were used to define the normal ranges for hTERT mRNA expression levels and gene copy number. Tissues were collected within 30 min after resection, snap-frozen and stored in liquid nitrogen until used; all were verified by histopathology. The collection of tissue and clinical and follow-up data was carried out in accordance with guidelines established by the Research Ethics Board (REB) of the University Health Network (UHN), which also approved this study.
DNA isolation and laser-captured microdissection
DNA was isolated from tumour cells micro-dissected using the Arcturus Pixcell II (Mountain View, CA, USA) laser capture microdissection (LCM) system. This includes 40 adenocarcinomas (ADC), 37 squamous cell carcinomas (SQCC), three adenosquamous carcinomas (ADSQC) and one large cell carcinoma (LCC). In addition, DNA was also extracted from 19 non-neoplastic lung samples. The tumour cells micro-dissected using LCM system were incubated in DNA extraction buffer containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.1 mg ml−1 gelatin, 0.45% Nonidet P-40, 0.45% Tween 20 and 0.4 mg ml−1 proteinase K. DNA was subsequently extracted by the phenol–chloroform method (Zhu et al, 2004).
Quantitative polymerase chain reaction
The quantitative polymerase chain reaction (qPCR) was performed using the SYBR Green technique in an ABI Prism 7700 sequence Detection System (Applied Biosystem, Foster City, CA, USA). The primer sequences were: hTERT sense 5′-taa aat tat cca cat ggc tca cgt-3′, antisense 5′-ctt ggg aac cag gac aaa gg-3′; PIK3R1 sense 5′-atc tgc cac tgg ctt ccc tt-3′, antisense 5′-cag tct ttc cct gat cat tga acc-3′. The PCR conditions were optimized as reported (Zhu et al, 2004). The PIK3R1 (5q13.1) gene is used as the reference nonamplified gene in NSCLC (Massion et al, 2002), and the hTERT gene copy number was estimated using comparative CT method. DNA from normal male lymphocyte (Novagen, San Diego, CA, USA) was used as the reference DNA. With this method, samples with normal copy number (disomy at both loci) or balanced polysomy (increased but equal copy number of both the reference and hTERT loci) will have an hTERT/PIK3R1 ratio of 1. Copy number values above or below the 2 standard deviations (s.d.) of mean of normal lung tissues values were designated as amplified or loss of the hTERT gene copy. Tumours with hTERT amount within the 2 s.d. of mean of normal lung tissues were classified as showing nonamplified samples.
Fluorescence in situ hybridisation
Archival paraffin blocks of 59 tumours that had been studied by qPCR were retrieved for fluorescence in situ hybridisation (FISH) analysis. Sections (4 μm) were mounted on positively charged slides and baked flat for 12–16 h at 56°C. Slides were dewaxed in three changes of xylene for 10 min each, followed by two changes in 100% ethanol for 5 min each. After air-drying, slides were treated in 2 × SSC for 20 min at 75°C, then for 5 min at room temperature (RT). The sections were then treated with 0.25 mg ml−1 proteinase K (Roche, Laval, QC, Canada) in 2 × SSC at 45°C for 20 min, followed by washing in 2 × SSC at RT for 5 min and serial dehydration through 70, 90 and 100% ethanol, and then left to air dry.
The hTERT/5q dual-colour FISH probe cocktail (Qbiogene, Montreal, QC, Canada) was applied at 0.02–0.06 μl mm−2 and sealed with rubber cement. The probe and target DNA were codenatured by heating to 80°C for 10 min in a Hybrite slide incubator (Vysis/Abbott Laboratories, Markham, ON, Canada). Hybridisation was for 16–20 h at 37°C in a moist light-sealed chamber in a dry oven. The slides were washed in two changes of 2 × SSC with 0.1% SDS at 45°C for 5 min each, followed by 5 min in 2 × SSC at RT. Slides were partially air-dried and 20–30 μl of DAPI mounting medium with antifade (Vector Labs, Burlingame, CA, USA) was applied, then cover slipped without sealing. Slides were stored in the dark at −20°C prior to imaging.
The FISH images were captured using the AxioImager system (Zeiss, Göttingen, Germany) with Z-stacking capabilities. Tumour cell nuclei identified using a DAPI filter and Z-stacked three-channel colour images (DAPI, FITC and Rhodamine/Cy3) were captured at × 63 or × 100 under oil immersion. Intact, nonoverlapping tumour cell nuclei (minimum 50 per case) without juxtaposed FISH signals were scored for the number of green (5p15.33) hTERT locus and red (5q31) control signals. The surrounding nontumour cells provide baseline estimation of the normal FISH signals (two green and two red signals). For survival analysis, high gene copy number cases included tumours with high polysomy (⩾4 hTERT gene copy in more than 40% of the tumour cells) or amplification (presence of tight hTERT gene clusters and a ratio of hTERT to chromosome of ⩾2 or ⩾15 copies of gene per cell in ⩾10% of analysed tumour cells), as defined by Cappuzzo et al (2005) for their study of the role of epidermal growth factor receptor (EGFR) gene copy number in EGFR inhibitor therapy.
Reverse transcription–qPCR
The mRNA expression was assayed using reverse transcription (RT)–qPCR on total RNA of 130 primary NSCLC and 18 corresponding non-neoplastic lung tissues using the ABI PRISM 7700 Sequence Detection System (Zhu et al, 2004). Total cellular RNA was isolated from the frozen tissues, as previously described (Tsao et al, 1998) and purified by the RNeasy Mini kit (Qiagen Inc., Mississauga, ON, Canada). The quality of the RNA preparations was confirmed by the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). In total, 5 μg of RNA was reverse transcribed using the Taqman reverse transcription reagent (Applied Biosystems, Branchburg, NJ, USA) in 100 μl reaction solution according to the manufacturer's instruction. After appropriate dilution, duplicate of 10 ng of cDNA was used as template for qPCR analysis of each sample. Primers were designed to span two adjacent exons to avoid amplification of contaminating genomic DNA sequences. The primers for hTERT were: sense 5′-cgtcgagctgctcaggtctt-3′, antisense 5′-agt gctgtctgattccaatgctt-3′. The ΔCT method was used to normalize the sample-to-sample variation in RNA/cDNA quantity using the 18s ribosomal RNA as the housekeeping gene (Zhu et al, 2004).
Statistical analysis
The Spearman correlation, χ2 tests or Fisher's exact test were used appropriately to assess association within and between molecular indices and the pathological or clinical factors. The end points for analyses were overall survival (from date of surgery to date of death) and recurrence-free survival (from date of surgery to date of recurrence). Cox proportional hazards regression was used in univariate and multivariate analyses. For Kaplan–Meier analysis, gene copy number and mRNA expression level were dichotomized using the upper limits of 95% confidence interval (95% CI) (mean+2 s.d.) for normal samples into nonamplified vs amplified or normal expression vs overexpression groups. Kaplan–Meier analysis estimates the survival of patient groups, and significant differences were determined by the log-rank test.
Results
Patient characteristics
Table 1 shows the demographics of patients in the studies of hTERT gene copy assessment by qPCR (n=81) or FISH (n=59), and hTERT mRNA expression (n=130) by RT–qPCR. There were no significant differences in the age, gender, stage and tumour differentiation grade among the three groups, but the mRNA expression study included more ADC patients. More than 90% of patients were stage I–II. The median follow-up was 3.19 (0.24–7.93) years, and 11 patients died without a relapse.
hTERT gene amplification
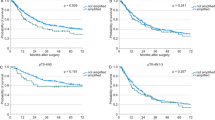
Figure 1A shows the distribution of relative gene copy of hTERT in normal and NSCLC. Using the upper limit of 95% CI (mean+2 s.d.) for normal samples as cutoff, hTERT gene amplification was found in 57% (46 of 81) of NSCLC patients. Gene copy loss was not observed. Amplification was more common in ADC compared to SQCC (Figure 1A), but was not correlated with tumour stage or differentiation grade (Table 2). The Kaplan–Meier survival estimation showed that patients with hTERT amplification by qPCR had poorer recurrence-free survival (log rank test P=0.02, Figure 2A). An analysis using 2 × mean of normal+2 s.d. as the cutoff to identify highly amplified patients showed statistically not significant separation of the survival curves of amplified vs unamplified patients, but further analysis showed that patients with gene copy changes between ⩾mean of normal+2 s.d. and <2 × mean of normal+2 s.d. showed similar survival outcome as the highly amplified (⩾mean of normal+2 s.d.) group, indicating that low amplification patients also experienced poorer survival outcome (Supplementary Figure 1). A similar trend of poorer overall survival for patients with amplified hTERT gene was noted, but this did not reach significance (log rank test P=0.15, Figure 2B).
Cox proportional hazards regression also showed a significant association of hTERT amplification with increased risk for death from recurrence (hazard ratio (HR) 2.16, 95% CI 1.07–4.37; P=0.03), but the correlation with poorer overall survival did not reach significance (HR 1.70, 95%CI 0.82–3.52; P=0.16). The hTERT amplification remained a significant prognostic marker for shorter recurrence-free survival (HR 2.06, 95%CI 1.01–4.2; P=0.05) after adjusting for the patient age, tumour stage and differentiation grade.
Because there was a high frequency (38%) of patients who were lost to follow-up at greater than 3 years after surgery, the 3-year survival rates were also estimated (Table 3). hTERT amplification was significantly associated with poorer recurrence-free survival (HR 2.96, 95% CI 1.27–6.90, P=0.01) and overall survival (HR 2.04, 95% CI 0.89–4.66, P=0.09) at 3 years. Multivariate analysis adjusting for patient age, tumour stage and differentiation grade confirmed that amplification was an independent prognostic marker for recurrence-free survival (HR 2.97, 95% CI 1.26–6.99, P=0.01; Table 3).
Validation of qPCR data with FISH
Fluorescence in situ hybridisation was performed on 59 tumours that had been studied for hTERT gene copy by qPCR (Figure 3). The ratios between the hTERT (green probe) and 5q13 reference gene locus D5S89 (red probe) signals were significantly correlated with the qPCR ratios of hTERT/PIK3R1 gene content (Spearman correlation coefficient r=0.43, P=0.0006). A better correlation was found for ADC (r=0.61, P=0.0003) than for SQCC (r=0.34, P=0.086). hTERT gene amplification by FISH was found in 73% (43 out of 59) of tumours and there were significant correlations between qPCR and FISH results for all tumours (P=0.008, Table 4) and for ADC (P=0.004). Although patients with high hTERT gene copy number (high polysomy and amplification) by FISH were more likely to experience early recurrence compared to those with lower gene copy numbers (low polysomy, trisomy or disomy), the difference was not statistically significant (HR 1.51, 95% CI 0.61–3.76, P=0.37).
Representative fluorescent in situ hybridisation (FISH) images. (A) A tumour with diploid genotype showing most tumour cell nuclei containing two green signals (hTERT, 5p15.33) and two red signals (control locus, 5q31). (B) A polysomy tumour showing several signals of both the hTERT and 5q; (C) Tumour with high-level amplification with their nuclei containing 10–30 hTERT signals and two or more 5q signals.
hTERT mRNA expression
Reverse transcription–qPCR did not detect hTERT mRNA expression in several non-neoplastic lung samples; therefore, the expression level of each sample was arbitrarily represented relative to the median of the entire data set. We used the mean+2 s.d. of the non-neoplastic lung expression levels as the cutoff to dichotomise tumours into hTERT normal expression and overexpression groups. Overexpression occurred in 72% (94 out of 130) of NSCLC, but was significantly more frequent in SQCC (87%) compared to ADC (68%) (Figure 1B and Table 2). Overexpression was also associated with higher tumour stages (Table 2). Among tumours with expression data, hTERT gene copy results by qPCR were also available for 67 cases (Table 5). Overexpression correlated with amplification (P=0.03) but only among the ADC (P=0.05). There were only 54 tumours with both FISH and expression results; high hTERT gene copy by FISH was not correlated with mRNA overexpression (data not shown).
Kaplan–Meier estimation revealed only trends for association between hTERT mRNA overexpression with recurrence-free survival or overall survival (log rank P=0.24 and P=0.13, respectively, Figure 2C and D), but it was significant for reduced overall survival at the 3-year follow-up time (log rank P=0.03, Table 3). Cox proportional hazards regression also showed that overexpression was an independent prognostic marker for overall survival at 3-year follow-up (HR 2.29, 95% CI 1.06–4.96, P=0.04) after adjusting for age, tumour stage and differentiation.
Discussion
We have evaluated the clinical and pathological significance of hTERT gene amplification and mRNA overexpression in NSCLC patients who were treated primarily by surgical resection. The hTERT gene amplification occurred in 57% of NSCLC, but this was more common among ADC (75%) than SQCC (35%). Among ADC, hTERT mRNA overexpression was significantly correlated with gene amplification (P=0.05). However, 87% (33 of 38) of SQCC also showed overexpression. These findings suggest that amplification is responsible for hTERT mRNA overexpression in a majority of ADC, while epigenetic factors at the transcriptional or post-transcriptional levels significantly affect hTERT expression. Most importantly, we have demonstrated that hTERT amplification is an independent prognostic marker for shorter recurrence-free survival in NSCLC patients.
Although many studies have examined the prognostic significance of hTERT mRNA/protein expression or activity in NSCLC (Table 6), to our knowledge, this is the first study that examined the prognostic value of hTERT gene amplification in lung cancer patients. The hTERT gene amplification was common in cell lines and primary tumours of lung, cervix, breast and in neuroblastoma (Zhang et al, 2000). Using ⩾5 copies of hTERT gene copy per nucleus in at least 20% of the cells to define amplification, Zhang et al (2000) reported hTERT amplification in 38% (eight out of 21) of lung carcinomas. Using qPCR that defines amplification as tumours with hTERT gene content greater than that of PIK3R1 (5q13.1), we found amplification in 57% of NSCLC. We also found a significant concordance between hTERT gene copy number assayed by qPCR and FISH (Spearman correlation coefficient r=0.43, P=0.0006), indicating that qPCR may serve as an alternative method to assay amplification.
There are some differences in the results of qPCR and FISH analyses on tumours. The qPCR assay cannot, whereas FISH can distinguish balanced copy gains (trisomy or polysomy) from diploid. To identify the former by qPCR, multiple reference genes or sequences on other chromosomes also need to be measured. The qPCR results also reflect the average gene copy number changes in DNA derived from thousands of microdissected tumour cells, whereas only a relatively small fraction (50 to few hundreds) of tumour cells are selected for FISH scoring. The poorer correlation between qPCR and FISH in SQCC compared to ADC could be due to the tendency of FISH to score tumour giant cells (cells with multilobated nuclei or multiple fused nuclei) that commonly contained multiple (5–10) copies of both hTERT and 5q reference loci; such giant tumour cells are more common in SQCC compared to ADC. Their scores may skew the assessment of gene copy number and introduce a higher variability in copy number estimation. Thus, the inability of our FISH studies to predict early recurrence of patients with hTERT gene amplification could result from: (1) smaller number of patients studied by FISH compared to qPCR, (2) cellular heterogeneity that skewed the overall FISH score due to recently acquired amplification in a discrete region within the tumour and (3) inconsistencies in FISH signal scoring criteria associated with polyploid nuclei and sectioning artefacts. Amplification has been defined differently in various FISH studies, such as ⩾3 (Murnane and Sabatier, 2004) or ⩾5 (Zhang et al, 2000) gene copies in at least 20% of the cells, or presence of tight gene clusters and a ratio of gene to chromosome of ⩾2 or ⩾15 copies of gene per cell in ⩾10% of analysed cells (Cappuzzo et al, 2005).
The reported frequencies of hTERT mRNA or protein overexpression in NSCLC ranged from 33 to 94%. In general, RT–qPCR data tended to demonstrate a higher overexpression rate compared to immunohistochemistry or mRNA in situ hybridization (mISH). Similar to our finding, most RT–qPCR studies but not other methods have also detected trace hTERT mRNA expression in normal lung tissues. We also found a correlation between hTERT amplification and overexpression, but mainly among the ADC.
The prognostic significance of hTERT expression or activity in NSCLC remains controversial (Table 6). Such an association has been reported in mISH (Kumaki et al, 2001; Wang et al, 2002; Fujita et al, 2003; Lantuejoul et al, 2004) and immunohistochemistry studies (Kumaki et al, 2001; Toomey et al, 2001; Lantuejoul et al, 2004). In contrast, the prognostic value of hTERT mRNA expression assayed by RT–qPCR has been inconsistent (Komiya et al, 2000; Hara et al, 2001; Marchetti et al, 2002; Wu et al, 2003; Hsu et al, 2004). Some studies have associated overexpression with poor prognosis (Komiya et al, 2000; Hara et al, 2001; Marchetti et al, 2002; Hsu et al, 2004), while others have failed to do so (Wu et al, 2003). One possible explanation is that hTERT mRNA has six known splice variants, but only the full-length transcript is functional. The qPCR probes designed in these studies did not distinguish between these variants. Although full-length mRNA level is usually proportional to the other variants (Fujiwara et al, 2004), it may range from 5 to 54% (Yi et al, 2001; Fujiwara et al, 2004). However, our finding that hTERT overexpression was predictive of poorer overall survival at 3 years is in agreement with a majority of these studies.
In conclusion, we have for the first time provided evidence that hTERT gene amplification or high copy number could be a marker for poorer prognosis in early-stage NSCLC patients, perhaps even more reliably than hTERT overexpression. While the clinical application of our findings requires more extensive retrospective and prospective validations in additional and larger cohorts of patients, further studies to evaluate the prognostic significance of hTERT by FISH is also warranted. In such case, a more refined FISH scoring system with reproducible criteria to identify clinically and biologically valid hTERT gene amplification would need to be developed. Since hTERT reactivation is a mechanism for cancer cells to avoid senescence (Shay and Roninson, 2004) and the latter could be induced by chemotherapy, the predictive value of hTERT amplification for benefit to adjuvant chemotherapy also needs evaluation (Winton et al, 2005). Recently, telomerase has been intensively studied as a target for novel cancer gene therapy and therapeutics (reviewed in Shay and Wright, 2002; Keith et al, 2004). Our finding that different types of NSCLC may alternately regulate hTERT overexpression suggests that patients with hTERT amplification could have different responses to telomerase-based therapies. The possible differential role of hTERT gene dosage in the diagnosis and treatment of lung cancer patients should be further investigated.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Albanell J, Lonardo F, Rusch V, Engelhardt M, Langenfeld J, Han W, Klimstra D, Venkatraman E, Moore MA, Dmitrovsky E (1997) High telomerase activity in primary lung cancers: association with increased cell proliferation rates and advanced pathologic stage. J Natl Cancer Inst 89: 1609–1615
Arinaga M, Shimizu S, Gotoh K, Haruki N, Takahashi T, Takahashi T, Mitsudomi T (2000) Expression of human telomerase subunit genes in primary lung cancer and its clinical significance. Ann Thorac Surg 70: 401–405; discussion 405–6
Bieche I, Nogues C, Paradis V, Olivi M, Bedossa P, Lidereau R, Vidaud M (2000) Quantitation of hTERT gene expression in sporadic breast tumors with a real-time reverse transcription–polymerase chain reaction assay. Clin Cancer Res 6: 452–459
Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn Jr PA, Varella-Garcia M (2005) Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 97: 643–655
Ducrest AL, Szutorisz H, Lingner J, Nabholz M (2002) Regulation of the human telomerase reverse transcriptase gene. Oncogene 21: 541–552
Fan X, Wang Y, Kratz J, Brat DJ, Robitaille Y, Moghrabi A, Perlman EJ, Dang CV, Burger PC, Eberhart CG (2003) hTERT gene amplification and increased mRNA expression in central nervous system embryonal tumors. Am J Pathol 162: 1763–1769
Fujita Y, Fujikane T, Fujiuchi S, Nishigaki Y, Yamazaki Y, Nagase A, Shimizu T, Ohsaki Y, Kikuchi K (2003) The diagnostic and prognostic relevance of human telomerase reverse transcriptase mRNA expression detected in situ in patients with nonsmall cell lung carcinoma. Cancer 98: 1008–1013
Fujiwara M, Kamma H, Wu W, Hamasaki M, Kaneko S, Horiguchi H, Matsui-Horiguchi M, Satoh H (2004) Expression and alternative splicing pattern of human telomerase reverse transcriptase in human lung cancer cells. Int J Oncol 24: 925–930
Hara H, Yamashita K, Shinada J, Yoshimura H, Kameya T (2001) Clinicopathologic significance of telomerase activity and hTERT mRNA expression in non-small cell lung cancer. Lung Cancer 34: 219–226
Harada K, Kurisu K, Arita K, Sadatomo T, Tahara H, Tahara E, Ide T, Uozumi T (1999) Telomerase activity in central nervous system malignant lymphoma. Cancer 86: 1050–1055
Hirashima T, Komiya T, Nitta T, Takada Y, Kobayashi M, Masuda N, Matui K, Takada M, Kikui M, Yasumitu T, Ohno A, Nakagawa K, Fukuoka M, Kawase I (2000) Prognostic significance of telomeric repeat length alterations in pathological stage I–IIIA non-small cell lung cancer. Anticancer Res 20: 2181–2187
Hsu CP, Miaw J, Shai SE, Chen CY (2004) Correlation between telomerase expression and terminal restriction fragment length ratio in non-small cell lung cancer – an adjusted measurement and its clinical significance. Eur J Cardiothorac Surg 26: 425–431
Kang SS, Kwon T, Kwon DY, Do SI (1999) Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J Biol Chem 274: 13085–13090
Keith WN, Bilsland A, Hardie M, Evans TRJ (2004) Drug insight: cancer cell immortality – telomerase as a target for novel cancer gene therapies. Nat Clin Prac Oncol 1: 88–96
Kolquist KA, Ellisen LW, Counter CM, Meyerson M, Tan LK, Weinberg RA, Haber DA, Gerald WL (1998) Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet 19: 182–186
Komiya T, Kawase I, Nitta T, Yasumitsu T, Kikui M, Fukuoka M, Nakagawa K, Hirashima T (2000) Prognostic significance of hTERT expression in non-small cell lung cancer. Int J Oncol 16: 1173–1177
Krams M, Hero B, Berthold F, Parwaresch R, Harms D, Rudolph P (2003) Full-length telomerase reverse transcriptase messenger RNA is an independent prognostic factor in neuroblastoma. Am J Pathol 162: 1019–1026
Kumaki F, Kawai T, Hiroi S, Shinomiya N, Ozeki Y, Ferrans VJ, Torikata C (2001) Telomerase activity and expression of human telomerase RNA component and human telomerase reverse transcriptase in lung carcinomas. Hum Pathol 32: 188–195
Lantuejoul S, Soria JC, Moro-Sibilot D, Morat L, Veyrenc S, Lorimier P, Brichon PY, Sabatier L, Brambilla C, Brambilla E (2004) Differential expression of telomerase reverse transcriptase (hTERT) in lung tumours. Br J Cancer 90: 1222–1229
Lee BK, Diebel E, Neukam FW, Wiltfang J, Ries J (2001) Diagnostic and prognostic relevance of expression of human telomerase subunits in oral cancer. Int J Oncol 19: 1063–1068
Lu C, Soria JC, Tang X, Xu X-C, Wang L, Mao L, Lotan R, Kemp B, Bekele BN, Feng L, Hong WK, Khuri FR (2004) Prognostic factors in resected stage I non-small cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol 22: 4575–4583 (abstract)
Luk C, Tsao MS, Bayani J, Shepherd F, Squire JA (2001) Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet 125: 87–99
Marchetti A, Pellegrini C, Buttitta F, Falleni M, Romagnoli S, Felicioni L, Barassi F, Salvatore S, Chella A, Angeletti CA, Roncalli M, Coggi G, Bosari S (2002) Prediction of survival in stage I lung carcinoma patients by telomerase function evaluation. Lab Invest 82: 729–736
Massion PP, Kuo WL, Stokoe D, Olshen AB, Treseler PA, Chin K, Chen C, Polikoff D, Jain AN, Pinkel D, Albertson DG, Jablons DM, Gray JW (2002) Genomic copy number analysis of non-small cell lung cancer using array comparative genomic hybridization: implications of the phosphatidylinositol 3-kinase pathway. Cancer Res 62: 3636–3640
Murnane JP, Sabatier L (2004) Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. BioEssays 26: 1164–1174
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959
Newbold RF (2002) The significance of telomerase activation and cellular immortalization in human cancer. Mutagenesis 17: 539–550
Ohali A, Avigad S, Cohen IJ, Meller I, Kollender Y, Issakov J, Gelernter I, Goshen Y, Yaniv I, Zaizov R (2003) Association between telomerase activity and outcome in patients with nonmetastatic Ewing family of tumors. J Clin Oncol 21: 3836–3843
Saretzki G, Petersen S, Petersen I, Kolble K, von Zglinicki T (2002) hTERT gene dosage correlates with telomerase activity in human lung cancer cell lines. Cancer Lett 176: 81–91
Shay JW, Roninson IB (2004) Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 23: 2919–2933
Shay JW, Wright WE (2002) Telomerase: a target for cancer therapeutics. Cancer Cell 2: 257–265
Taga S, Osaki T, Ohgami A, Imoto H, Yasumoto K (1999) Prognostic impact of telomerase activity in non-small cell lung cancers. Ann Surg 230: 715–720
Tchirkov A, Rolhion C, Kemeny JL, Irthum B, Puget S, Khalil T, Chinot O, Kwiatkowski F, Perissel B, Vago P, Verrelle P (2003) Clinical implications of quantitative real-time RT–PCR analysis of hTERT gene expression in human gliomas. Br J Cancer 88: 516–520
Toomey D, Smyth G, Condron C, Kay E, Conroy R, Foley D, Hong C, Hogan B, Toner S, McCormick P, Broe P, Kelly C, Bouchier-Hayes D (2001) Immune function, telomerase, and angiogenesis in patients with primary, operable nonsmall cell lung carcinoma: tumor size and lymph node status remain the most important prognostic features. Cancer 92: 2648–2657
Tsao M-S, Liu N, Chen J-R, Pappas J, Ho J, To C, Viallet J, Park M, Zhu H (1998) Differential expression of Met/hepatocyte growth factor receptor in subtypes of non-small cell lung cancer. Lung Cancer 20: 1–16
Vaziri H, Benchimol S (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 8: 279–282
Wang L, Soria JC, Kemp BL, Liu DD, Mao L, Khuri FR (2002) hTERT expression is a prognostic factor of survival in patients with stage I non-small cell lung cancer. Clin Cancer Res 8: 2883–2889
Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, Cormier Y, Goss G, Inculet R, Vallieres E, Fry W, Bethune D, Ayoub J, Ding K, Seymour L, Graham B, Tsao M-S, Gandara D, Kesler K, Demmy T, Shepherd F (2005) Vinorelbine plus cisplatin vs observation in resected non-small-cell lung cancer. N Engl J Med 353: 2589–2597
Wu TC, Lin P, Hsu CP, Huang YJ, Chen CY, Chung WC, Lee H, Ko JL (2003) Loss of telomerase activity may be a potential favorable prognostic marker in lung carcinomas. Lung Cancer 41: 163–169
Yi X, Shay JW, Wright WE (2001) Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 29: 4818–4825
Zhang A, Zheng C, Hou M, Lindvall C, Wallin KL, Angstrom T, Yang X, Hellstrom AC, Blennow E, Bjorkholm M, Zetterberg A, Gruber A, Xu D (2002) Amplification of the telomerase reverse transcriptase (hTERT) gene in cervical carcinomas. Genes Chromosomes Cancer 34: 269–275
Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, Yan Z, Yang X, Henriksson M, Blennow E, Nordenskjold M, Zetterberg A, Bjorkholm M, Gruber A, Xu D (2000) Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res 60: 6230–6235
Zhu CQ, Blackhall FH, Pintilie M, Iyengar P, Liu N, Ho J, Chomiak T, Lau D, Winton T, Shepherd FA, Tsao MS (2004) Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker. Clin Cancer Res 10: 1984–1991
Acknowledgements
This work was supported by Grant # 015184 from the Canadian Cancer Society through funding by the National Cancer Institute of Canada. Dr Tsao holds the M Qasim Choksi Chair in Lung Cancer Translational Research. Dr Squire holds the JC Boileau Grant Chair in Oncologic Pathology. Dr Shepherd holds the Scott Taylor Chair in Lung Cancer Research. Dr Cutz was a Fellow in the CIHR Training Programme (STP-53912) for Clinician Scientist in Molecular Oncologic Pathology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Zhu, CQ., Cutz, JC., Liu, N. et al. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br J Cancer 94, 1452–1459 (2006). https://doi.org/10.1038/sj.bjc.6603110
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6603110
Keywords
This article is cited by
-
Assessment of MYC and TERT copy number variations in lung cancer using digital PCR
BMC Research Notes (2023)
-
Connecting telomere maintenance and regulation to the developmental origin and differentiation states of neuroblastoma tumor cells
Journal of Hematology & Oncology (2022)
-
A pan-cancer landscape of telomeric content shows that RAD21 and HGF alterations are associated with longer telomeres
Genome Medicine (2022)
-
TERT amplification but not activation of canonical Wnt/β-catenin pathway is involved in acral lentiginous melanoma progression to metastasis
Modern Pathology (2020)
-
Strong antigen-specific T-cell immunity induced by a recombinant human TERT measles virus vaccine and amplified by a DNA/viral vector prime boost in IFNAR/CD46 mice
Cancer Immunology, Immunotherapy (2019)