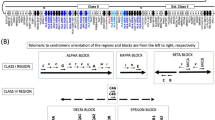
Abstract
The cytotoxic T lymphocyte antigen 4 (CTLA4) acts as a potent negative regulator of T-cell response, and has been suggested as a pivotal candidate gene for autoimmune disorders such as Graves' disease, type 1 diabetes and autoimmune hypothyroidism, among others. Several single-nucleotide polymorphisms (SNPs) have been proposed as the susceptibility variants, or to be in strong linkage disequilibrium (LD) with the variant. Nevertheless, contradictory results have been found, which may be due to lack of knowledge of the genetic structure of CTLA4 and its geographic variation. We have typed 17 SNPs throughout the CTLA4 gene region in order to analyze the haplotype diversity and LD structure in a worldwide population set (1262 individuals from 44 populations) to understand the variation pattern of the region. Allele and haplotype frequency differentiation between populations is consistent with genomewide averages and points to a lack of strong population-specific selection pressures. LD is high and its pattern is not significantly different within or between continents. However, haplotype composition is significantly different between geographical groups. A continent-specific set of haplotype tagging SNPs has been designed to be used for future association studies. These are portable among populations, although their efficiency might vary depending on the population haplotype spectrum.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Walunas TL, Lenschow DJ, Bakker CY et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994; 1: 405–413.
Walunas TL, Bakker CY, Bluestone JA . CTLA-4 ligation blocks CD28-dependent T cell activation. J Exp Med 1996; 183: 2541–2550.
Khattri R, Auger JA, Griffin MD, Sharpe AH, Bluestone JA . Lymphoproliferative disorder in CTLA-4 knockout mice is characterized by CD28-regulated activation of Th2 responses. J Immunol 1999; 162: 5784–5791.
Harper K, Balzano C, Rouvier E, Mattei MG, Luciani MF, Golstein P . CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J Immunol 1991; 147: 1037–1044.
Martin AM, Athanasiadis G, Greshock JD et al. Population frequencies of single nucleotide polymorphisms (SNPs) in immuno-modulatory genes. Hum Hered 2003; 55: 171–178.
Johnson GC, Esposito L, Barratt BJ et al. Haplotype tagging for the identification of common disease genes. Nat Genet 2001; 29: 233–237.
Ueda H, Howson JM, Esposito L et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003; 423: 506–511.
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K . A comprehensive review of genetic association studies. Genet Med 2002; 4: 45–61.
Ide A, Kawasaki E, Abiru N et al. Association between IL-18 gene promoter polymorphisms and CTLA-4 gene 49A/G polymorphism in Japanese patients with type 1 diabetes. J Autoimmun 2004; 22 (1): 73–78.
Lee CS, Lee YJ, Liu HF et al. Association of CTLA4 gene A–G polymorphism with rheumatoid arthritis in Chinese. Clin Rheumatol 2003; 22: 221–224.
Teutsch SM, Booth DR, Bennetts BH, Heard RN, Stewart GJ . Association of common T cell activation gene polymorphisms with multiple sclerosis in Australian patients. J Neuroimmunol 2004; 148: 218–230.
Vaidya B, Oakes EJ, Imrie H et al. CTLA4 gene and Graves' disease: association of Graves' disease with the CTLA4 exon 1 and intron 1 polymorphisms, but not with the promoter polymorphism. Clin Endocrinol (Oxford) 2003; 58: 732–735.
van Oosterhout AJ, Deurloo DT, Groot PC . Cytotoxic T lymphocyte antigen 4 polymorphisms and allergic asthma. Clin Exp Allergy 2004; 34: 4–8.
Hudson LL, Silver RM, Pandey JP . Ethnic differences in cytotoxic T lymphocyte associated antigen 4 genotype associations with systemic sclerosis. J Rheumatol 2004; 31: 85–87.
Hudson LL, Rocca K, Song YW, Pandey JP . CTLA-4 gene polymorphisms in systemic lupus erythematosus: a highly significant association with a determinant in the promoter region. Hum Genet 2002; 111: 452–455.
Fernandez-Blanco L, Perez-Pampin E, Gomez-Reino JJ, Gonzalez A . A CTLA-4 polymorphism associated with susceptibility to systemic lupus erythematosus. Arthritis Rheum 2004; 50: 328–329.
Bouqbis L, Izaabel H, Akhayat O et al. Association of the CTLA4 promoter region (−1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun 2003; 4: 132–137.
Cardon LR, Bell JI . Association study designs for complex diseases. Nat Rev Genet 2001; 2: 91–99.
Bertranpetit J, Calafell F, Comas D, Gonzalez-Neira A, Navarro A . Structure of linkage disequilibrium in humans: genome factors and population stratification. Cold Spring Harb Symp Quant Biol 2003; 68: 79–88.
Tishkoff SA, Dietzsch E, Speed W et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science 1996; 271: 1380–1387.
Sawyer SL, Mukherjee N, Pakstis AJ et al. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet 2005; 13: 677–686.
Gonzalez-Neira A, Calafell F, Navarro A et al. Geographic stratification of linkage disequilibrium: a worldwide population study in a region of chromosome 22. Hum Genomics 2004; 1: 399–409.
Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES . High-resolution haplotype structure in the human genome. Nat Genet 2001; 29: 229–232.
Wall JD, Pritchard JK . Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet 2003; 4: 587–597.
Gabriel SB, Schaffner SF, Nguyen H et al. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Clark AG, Weiss KM, Nickerson DA et al. Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 1998; 63: 595–612.
Wang N, Akey JM, Zhang K, Chakraborty R, Jin L . Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 2002; 71: 1227–1234.
Weale ME, Depondt C, Macdonald SJ et al. Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet 2003; 73: 551–565.
Mueller JC, Lohmussaar E, Magi R et al. Linkage disequilibrium patterns and tagSNP transferability among European populations. Am J Hum Genet 2005; 76: 387–398.
Crawford DC, Carlson CS, Rieder MJ et al. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet 2004; 74: 610–622.
Chen FC, Li WH . Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 2001; 68: 444–456.
Barbujani G, Magagni A, Minch E, Cavalli-Sforza LL . An apportionment of human DNA diversity. Proc Natl Acad Sci USA 1997; 94: 4516–4519.
Rosenberg NA, Pritchard JK, Weber JL et al. Genetic structure of human populations. Science 2002; 298: 2381–2385.
Excoffier L, Hamilton G . Comment on ‘Genetic structure of human populations’. Science 2003; 300: 1877; author reply 1877.
Akey JM, Zhang G, Zhang K, Jin L, Shriver MD . Interrogating a high-density SNP map for signatures of natural selection. Genome Res 2002; 12: 1805–1814.
Kidd KK, Pakstis AJ, Speed WC, Kidd JR . Understanding human DNA sequence variation. J Hered 2004; 95: 406–420.
Tishkoff SA, Varkonyi R, Cahinhinan N et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 2001; 293: 455–462.
Hamblin MT, Thompson EE, Di Rienzo A . Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 2002; 70: 369–383.
Sabeti PC, Reich DE, Higgins JM et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002; 419: 832–837.
Kong A, Gudbjartsson DF, Sainz J et al. A high-resolution recombination map of the human genome. Nat Genet 2002; 31: 241–247.
Kidd JR, Pakstis AJ, Zhao H et al. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 2000; 66: 1882–1899.
Mateu E, Perez-Lezaun A, Martinez-Arias R et al. PKLR- GBA region shows almost complete linkage disequilibrium over 70 kb in a set of worldwide populations. Hum Genet 2002; 110: 532–544.
Kidd KK, Morar B, Castiglione CM et al. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet 1998; 103: 211–227.
Tishkoff SA, Goldman A, Calafell F et al. A global haplotype analysis of the myotonic dystrophy locus: implications for the evolution of modern humans and for the origin of myotonic dystrophy mutations. Am J Hum Genet 1998; 62: 1389–1402.
Mateu E, Calafell F, Lao O et al. Worldwide genetic analysis of the CFTR region. Am J Hum Genet 2001; 68: 103–117.
Gonzalez-Neira A, Ke X, Lao O et al. The portability of tagSNPs across populations. A worldwide survey (submitted).
Cann HM, de Toma C, Cazes L et al. A human genome diversity cell line panel. Science 2002; 296: 261–262.
Iyengar S, Seaman M, Deinard AS et al. Analyses of cross species polymerase chain reaction products to infer the ancestral state of human polymorphisms. DNA Seq 1998; 8: 317–327.
Stephens M, Donnelly P . A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 2003; 73: 1162–1169.
Schneider S, Roessli D, Excoffier L . Arlequin Ver. 2.0: A software for Population Genetic Data Analysis, 2nd edn., University of Geneva, Geneva, 2000.
Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF . Minimal haplotype tagging. Proc Natl Acad Sci USA 2003; 100: 9900–9905.
Acknowledgements
We thank Elena Bosch, Arcadi Navarro, Michelle Gardner, Lourdes Sampietro and Mònica Vallés (Universitat Pompeu Fabra) for helpful advise and technical support. Mark Shriver (Pennsylvania State University) and Kenneth K Kidd (Yale University) kindly provided the raw data for the FST comparison. We thank Howard Cann (CEPH, Paris) for providing the HGDP-CEPH panel. We also thank Anna Pérez-Lezaun, Roger Anglada and Stéphanie Plaza (Servei de Genòmica, Universitat Pompeu Fabra) for technical support. This research was supported by Ministerio de Educación y Ciencia of the Spanish Government (BFU2004-04208/BMC) and Departament d'Universitats, Recerca i Societat de la Informació (DURSI) of the Generalitat de Catalunya.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramírez-Soriano, A., Lao, O., Soldevila, M. et al. Haplotype tagging efficiency in worldwide populations in CTLA4 gene. Genes Immun 6, 646–657 (2005). https://doi.org/10.1038/sj.gene.6364251
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6364251
Keywords
This article is cited by
-
The effect of CTLA-4 A49G polymorphism on rheumatoid arthritis risk: a meta-analysis
Diagnostic Pathology (2014)
-
Family-based association study of cytotoxic T-lymphocyte antigen-4 with susceptibility to Graves' disease in Han population of Taiwan
Genes & Immunity (2008)
-
Comparison of ENCODE region SNPs between Cebu Filipino and Asian HapMap samples
Journal of Human Genetics (2007)
-
Transferability of tag SNPs in genetic association studies in multiple populations
Nature Genetics (2006)