Abstract
Hypotonia–cystinuria syndrome (HCS) is a recessive disorder caused by microdeletions of SLC3A1 and PREPL on chromosome 2p21. Patients present with generalized hypotonia at birth, failure to thrive, growth retardation and cystinuria type I. While the initially described HCS families live in small regions in Belgium and France, we have now identified HCS alleles in patients and carriers from the Netherlands, Italy, Canada and United States of America. Surprisingly, among the nine deletions detected in those patients, only one novel deletion was found. Furthermore, one previously described deletion was found six times, another twice. Finally, we have investigated the frequency of both deletions using a random Belgian cohort. Given the global occurrence, HCS should be considered in the differential diagnosis of neonatal hypotonia.
Similar content being viewed by others
Introduction
The hypotonia–cystinuria syndrome (HCS; MIM 606407) is a recessive congenital disorder characterized by generalized hypotonia at birth, failure to thrive, growth retardation and cystinuria type I.1, 2 Recently, the genetic defect underlying this syndrome has been characterized in nine families.1 The disease is due to microdeletions that disrupt two genes on chromosome 2p21, SLC3A1 and PREPL. As mutations in SLC3A1 cause isolated cystinuria type I and the flanking genes are normally expressed, the extended phenotype can be attributed to PREPL.1
A larger deletion of 179 kb at this locus has previously been reported,3, 4 which causes the recessive 2p21 deletion syndrome. At least four genes (SLC3A1, PREPL, PPM1B and C2orf34) are deleted, resulting in a much more severe phenotype. A detailed comparison between the two syndromes has been published.5
At least three additional deletions involving PREPL have been reported in patients with isolated cystinuria type I. However, they were not fully characterized.6, 7, 8 Finally, one sporadic case with HCS symptoms, accompanied with a mitochondrial respiratory chain defect has been reported. Molecular analysis of the SLC3A1/PREPL locus was suggested but not performed.9
This report focuses on the molecular analysis of four new HCS families, including two previously described cases. Finally, the frequency of the most prevalent deletions causing HCS was estimated using a random Belgian population.
Materials and methods
Patient material
Material from family 10 (University Hospitals Leuven, Belgium), family 11 (University Medical Centre Leiden, The Netherlands) and family 12 (CentraCare Clinic, St Cloud, MN, USA) was sent for molecular diagnosis for the SLC3A1/PREPL locus. DNA samples for family 13 (University of Verona, Italy) and family 14 (McGill University, Montreal, Canada) were obtained upon request. As a random population, we used the CAREGENE10 cohort.
Quantitative PCR
Quantitative PCR was performed using qPCR Master Mix for SYBR Green I detection and fluorescein as internal standard (Eurogentec, Seraing, Belgium) on the MyIQ system (Bio-Rad, Nazareth-Eke, Belgium), in accordance with the manufacturer's guidelines. Primers were developed with Primer Express software (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). The relative number of alleles were calculated using the Livak method.11 An amplicon within the p53 gene was used as reference.
Junction fragment PCR and sequencing
PCR products were amplified using 150 ng of genomic DNA, by the use of the Advantage 2 PCR kit (Clontech, Mountain View, CA, USA). Junction fragments were purified with the Montage PCR Centrifugal Filter Device (Millipore, Billerica, MA, USA) before sequencing (Bigdye Terminator v3.1, Applied Biosystems) on the ABI3130 system (Applied Biosystems). Primers used for amplifying deletions A and B have been described.1 Primers for deletion E are 5′-TTGCTCCAAAAGTTCCTAACCAA-3′ and 5′-TTCTGTGTTGAGGTTGCACTCC-3′.
Haplotype analysis
Genotypes of microsatellites and single nucleotide polymorphisms (SNP) were determined as described.1
Results
The clinical and endocrinological data on the patients are similar to those reported earlier.1 Details are listed in Table 1. In summary, the clinical picture is characterized by neonatal and infantile hypotonia, and anorexia, often necessitating nasogastric tube feeding or gastrostomy. Some dysmorphy is present, particularly dolichocephaly and ptosis of the eyelids, as well as a striking nasal speech. Anorexia and hypotonia ameliorate with age, but gradually growth velocity decreases due to IGF-1 insufficiency. Patients respond well to growth hormone treatment. Electromyography and brain magnetic resonance imaging are normal, when performed. In addition, patients have classical cystinuria type I causing nephrolithiasis at variable ages.
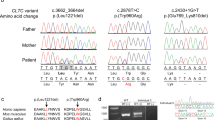
The presence or absence of different exons of SLC3A1 and PREPL was determined in patients and parents (when available) using quantitative PCR. In families 11 and 13, the boundaries for both deleted alleles were located in intron 1 of SLC3A1 and intron 1 of PREPL. As the same fragments were deleted in the previously described deletion B,1 we performed a junction fragment PCR specific for this deletion followed by sequencing of the amplicons (Figure 1). Both patients are homozygous for deletion B, therefore confirming the diagnosis of HCS. The patient in family 10 is compound heterozygous for two deletions, one identical to deletion B, the other extending from exon 10 in SLC3A1 to exon 2 in PREPL-like deletion A from Jaeken et al.1 Junction fragment PCR and sequencing confirmed that the second allele is indeed deletion A (Figure 1). The patient in family 12 showed a homozygous loss from exon 10 in SLC3A1 to exon 2 in PREPL. Junction fragment PCR and sequencing confirmed that the paternal allele is again deletion A. The proximal boundary of the deletion on the maternal chromosome is located in intron 1 of SLC3A1, whereas the distal break point is located between 45 and 50 kb upstream of PREPL (Figures 1 and 2). Sequencing of the junction fragment indicated that the deletion was 127 kb in size, with the distal break point located in intron 3 of C2orf34, the gene flanking PREPL. This is a novel allele (deletion E) involved in HCS.
Genetic analysis of the HCS patients. (A) Quantitative PCR analysis on genomic DNA of HCS patients. Only relevant amplicons are shown. (B) Junction fragments spanning the break point in patients and parents (when available). −, negative control; P, patient; M, mother; F, father. (C) Genotype of the patients.
The 2p21 locus. (a) Genomic organization of the 2p21 locus. The flanking microsatellite markers used in the haplotype analysis are indicated with their genomic coordinates. (b) Schematic representation of the deletions detected in the novel HCS families, presented here. Deletions (a and b) have previously been described,1 whereas deletion E represents a new allele. Sequences flanking the break points are shown and are numbered in accordance with the numbering in the database (Homo sapiens, build 36; http://www.ncbi.nlm.nih.gov/).
The patient in family 14 (patient 936 in Saadi et al6) has isolated cystinuria type I, and has previously been described as compound heterozygous for two deletions involving SLC3A1. The deletion on the paternal allele had been shown to extend from exon 1 to exon 7 of SLC3A1, whereas the deletion on the maternal allele starts in intron 1 of SLC3A1 and extends beyond exon 10. We have now determined the exact break points on the latter deletion. Quantitative PCR analysis revealed that the proximal boundary was located in intron 1 of SLC3A1, whereas the distal break point was localized in intron 2 of PREPL. Again, this pointed toward the presence of deletion B (Figure 1).
The different deletion alleles were haplotyped using microsatellites and SNPs. Only family 10 showed a partially conserved ancestral haplotype for deletion A (Figure 3). SNPs genotyped in families 11–13 identified the same alleles as present in the ancestral haplotype of deletion B; however, the genotypes for families 11 and 13 were not informative. The SNP data for family 10 could not be obtained due to a lack of parental DNA (data not shown).
Because of the high incidence of deletions A and B, we have screened a random Belgian cohort for the presence of these deletions. We found 1 allele with deletion A and 1 allele with deletion B in a total of 936 random samples (1872 chromosomes). This leads to an allele frequency estimate for deletions A or B between 1/333 and 1/7700 (binomial distribution; 95% confidence interval).
Discussion
HCS was initially described in nine families living in Belgium and France. On 18 independent chromosomes, only four different deletions (deletions A–D) were found. The frequency of deletions A and B was 4/18 and 11/18, respectively, due to founder effects.1 In the four novel HCS families and the one carrier of a HCS allele described here, the frequency of deletions A and B were 2/9 and 6/9, respectively. The allele frequencies of deletions A or B in a random Belgian cohort were estimated between 1/333 and 1/7700.
Deletions A and B are globally distributed, as they were found in patients from The Netherlands, Italy, Canada and the United States America. However, the ancestral haplotypes of the microsatellite markers flanking the break points were completely decayed in these families, indicating that the deletions are old. However, it cannot be excluded that a genetic mechanism is promoting deletions in this region. If this is the case, however, it is remarkable that the break points are exactly the same.
The patient in family 12 appeared to be compound heterozygous for deletions A and E, the latter being a novel deletion involved in HCS. This deletion does not only disrupt the coding region of SLC3A1 and PREPL, but also of C2orf34. C2orf34 has a putative methyltransferase domain, but its function has not yet been described. It is, however, not expected that a loss of one allele is modulating the phenotype in this HCS patient, because the mother showed no phenotype that might be due to the loss of one allele of C2orf34.
Unexpected results were obtained for family 13. The proband in this family showed all HCS symptoms, accompanied with a mitochondrial respiratory chain defect. As a mitochondrial respiratory chain defect was also observed in the 2p21 deletion syndrome,3 one might have expected that besides SLC3A1 and PREPL, at least one other gene (PPM1B or C2orf133) was involved. However, this patient is homozygous for deletion B, and therefore her genotype is identical to some of the previously published patients. She has been the only HCS patient with a reported respiratory chain defect. This brings up an intriguing question about the cause of the respiratory chain defect in this patient. A possible explanation could be the presence of variations in modifier genes, either in cis or in trans, yet to be discovered. Currently, in-depth analysis of mitochondrial respiratory chain function in fibroblast cells from other HCS patients is being performed to determine potential minor defects that might previously have been overlooked.
Three deletions involving PREPL have been described in patients with isolated cystinuria type I. We characterized the deletion initially reported in Saadi et al.6 It turns out to be the most frequent HCS allele, deletion B, only disrupting the coding region of SLC3A1 and PREPL. This leaves the total of unique alleles where PREPL is deleted at six: five deletions present in HCS and one causing the 2p21 deletion syndrome. Possibly, two other alleles exist, but the break points have not been determined.7, 8
In conclusion, HCS is a relatively homogenous syndrome, with common ancestral alleles found in different populations. On the basis of the allele frequencies, this disease will be rare (1/1 000 000 for an allele frequency of 1/1000); hence, our observations are compatible with a founder effect in the regions from where the HCS patients originate.
References
Jaeken J, Martens K, Francois I et al: Deletion of PREPL, a gene encoding a putative serine oligopeptidase, in patients with hypotonia–cystinuria syndrome. Am J Hum Genet 2006; 78: 38–51.
Clara R, Lowenthal A : Familial aminoaciduria with muscular hypotonia and dwarfism. Bull Acad R Med Belg 1966; 6: 577–611.
Parvari R, Brodyansky I, Elpeleg O, Moses S, Landau D, Hershkovitz E : A recessive contiguous gene deletion of chromosome 2p16 associated with cystinuria and a mitochondrial disease. Am J Hum Genet 2001; 69: 869–875.
Parvari R, Gonen Y, Alshafee I, Buriakovsky S, Regev K, Hershkovitz E : The 2p21 deletion syndrome: characterization of the transcription content. Genomics 2005; 86: 195–211.
Martens K, Derua R, Meulemans S et al: PREPL: a putative novel oligopeptidase propelled into the limelight. Biol Chem 2006; 387: 879–883.
Saadi I, Chen XZ, Hediger M et al: Molecular genetics of cystinuria: mutation analysis of SLC3A1 and evidence for another gene in type I (silent) phenotype. Kidney Int 1998; 54: 48–55.
Font-Llitjos M, Jimenez-Vidal M, Bisceglia L et al: New insights into cystinuria: 40 new mutations, genotype–phenotype correlation, and digenic inheritance causing partial phenotype. J Med Genet 2005; 42: 58–68.
Bisceglia L, Calonge MJ, Dello Strologo L et al: Molecular analysis of the cystinuria disease gene: identification of four new mutations, one large deletion, and one polymorphism. Hum Genet 1996; 98: 447–451.
Zaffanello M, Beghini R, Zamboni G, Fanos V : A sporadic case of cystinuria, respiratory chain and growth hormone deficiencies. Pediatr Nephrol 2003; 18: 846–847.
Defoor J, Martens K, Matthijs G et al: The care gene study: muscle-specific creatine kinase gene and aerobic power in coronary artery disease. Eur J Cardiovasc Prev Rehabil 2005; 12: 415–417.
Livak KJ, Schmittgen TD : Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001; 25: 402–408.
Acknowledgements
DNA from family 14 was kindly provided by Dr Rozen (McGill University, Montreal, Canada). We thank Dr Vos (Medical Centre Haaglanden, Den Haag, The Netherlands) for assistance with patient 11. Grant and scholarship support was provided by the ‘Vlaams Instituut voor de Bevordering van Wetenschappelijk-Technologisch Onderzoek in de Industrie (IWT)’ and the ‘Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO)’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martens, K., Heulens, I., Meulemans, S. et al. Global distribution of the most prevalent deletions causing hypotonia–cystinuria syndrome. Eur J Hum Genet 15, 1029–1033 (2007). https://doi.org/10.1038/sj.ejhg.5201881
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201881
Keywords
This article is cited by
-
The second point mutation in PREPL: a case report and literature review
Journal of Human Genetics (2018)
-
First cardiac manifestation of hypotonia-cystinuria syndrome
Metabolic Brain Disease (2018)
-
Cystinuria: an inborn cause of urolithiasis
Orphanet Journal of Rare Diseases (2012)
-
Clinical utility gene card for: Cystinuria
European Journal of Human Genetics (2012)
-
Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin
Nature Communications (2010)