Abstract
Hearing loss is the most common sensory disorder in humans, and genetic factors are a major cause. Approximately 15–20% of genetic cases exhibit an autosomal dominant pattern of transmission. So far, 41 autosomal dominant loci have been mapped and 17 genes have been identified. Here we report the mapping of a novel locus for autosomal dominant non-syndromic hearing loss, DFNA47, to chromosome 9p21–22 in a large multigenerational Italian family with progressive hearing impairment. Most affected individuals noticed hearing impairment after their teens with subsequent gradual progression to a moderate-severe loss. There were no obvious vestibular dysfunction and other associated abnormalities. A maximum lod score of 3.14 was obtained with marker D9S157 (at θ=0) after a genome wide search. The study of additional markers allowed us to confirm this region with positive lod scores of 3.58 (at θ=0 from D9S285) and of 3.67 (at θ=0 from D9S162). Recombinants define a region of approximately 9 cM flanked by markers D9S268 and D9S942. Multipoint linkage analysis showed a Lod score of 4.26. Few known genes map to the region, and those possibly related by function to hearing are being screened for disease-causing mutations.
Similar content being viewed by others
Introduction
Severe deafness or hearing impairment is the most prevalent inherited sensory disorder, affecting about 1 in 1000 children.1 In addition a large proportion of older adults suffer from progressive hearing loss.2 Most deafness results from peripheral auditory defects that occur as a consequence of either conductive (outer or middle ear) or sensorineuronal (cochlea) abnormalities.3 Nonsyndromic hearing loss is the most common form of genetic deafness and is a very heterogeneous trait with more than 70 loci so far reported4 (see for updated data the Hereditary Hearing Loss Homepage at the following URL: www.uia.ac.be/dnalab/hh/). A proportion comprised between 15 and 20% of cases is caused by autosomal dominant defects (Non Syndromic Autosomal Dominant or NSAD). Usually, hearing impairment is progressive, involving first high frequencies and then middle and lower frequencies, affects both ears (bilateral) and the degree of severity is comprised between mild to moderate, but in some cases severe to profound. To date 41 different NSAD hearing loss loci have been described and 17 different genes identified (see Hereditary Hearing Loss Homepage). This extreme genetic heterogeneity suggests that there are many different processes within the inner ear that, once altered, can cause hearing loss.
Materials and methods
Patients and DNA samples
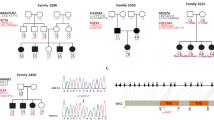
We included a large Italian kindred affected by NSAD hearing loss (Figure 1a) in a genome wide search. The family, coming from Southern Italy, is affected by progressive postlingual sensorineural deafness due to a mutation which appears to be completely penetrant, with a variable age of onset (30–35 years old for the first symptoms; 20–25 years old for the first audiometric abnormalities). Affected individuals experience high frequency hearing loss which progressed with age to include middle and lower frequencies. Around the age of 50 they invariably present a moderate to severe sensorineural deafness. Audiograms showed different degrees of hearing impairment, ranging from moderate to severe sensorineural hearing loss (data not shown). Tympanometry results indicated proper functioning of the tympanic membrane and middle ear (data not shown). Vestibular and/or visual involvement was excluded in all affected individuals either by physical examination and history. Peripheral blood was obtained from all subjects, and DNA was isolated from blood leukocytes according to standard methods.
Drawing of the large Italian multigenerational pedigree affected by non syndromic autosomal dominant hearing loss. The pedigree segregates with DFNA47. Black symbols represent individuals with hearing impairment due to DFNA47. Clear symbols represent healthy individuals. Haplotypes for the most closely linked microsatellites are shown below each symbol. Arrows mark recombinational events. Question mark on individual symbol means phenotype unknown.
Linkage analysis
The genome wide search was performed using the ABI PRISM Linkage Mapping Set v.2 (Perkin Elmer, USA), characterized by over 375 markers that define a 10 cM resolution human index map. PCR reactions using fluorescently labelled primers were run under the conditions suggested by the supplier. An aliquot of each PCR reaction was run on an ABI PRISM 3100 DNA sequencer and results were processed by GENESCAN software. Allele assignation was carried out using the Genotyper™ software. Statistical analysis was performed on the basis of an autosomal disease with complete penetrance. Individuals 23 and 30 are younger than the mean age of onset (18 and 20 years old, respectively) and we do not have any clinical and/or instrumental record of them because they refused to undergo any specific examination. Thus, their phenotype was considered as unknown. The disease-gene frequency was set to 0.012, and all marker alleles were considered to be equally frequent. We have performed a simulation study using SLINK and obtained an average maximum LOD score of 2.74 with this pedigree. Pairwise linkage analysis was performed using the MLINK program version 5.2 from the LINKAGE computer package5 and using FASTLINK version 4.1P.6 The approximate 95% confidence limits for the maximum recombination fraction (Thetamax) at the maximum LOD score (Zmax) were calculated by the 1-LOD-down method.5 Alleles were down-coded without loss of informativity to reduce computing time. The multipoint parametric analysis was performed by SimWalk2 v.2.82 using Markov chain MonteCarlo (MCMC) and simulated annealing algorithms.7 Files for SimWalk analysis were prepared using the Mega2 software.8
Results
A genome wide search to map the disease locus in this large Italian kindred has been performed using commercially available microsatellite panels. Pairwise linkage analysis showed a maximum Lod score of 3.58 with marker D9S285 at a recombination frequency of θ=0. Additional significant lod scores have been obtained with marker D9S157 (3.14) located centromeric to D9S162 (Table 1). Negative results were obtained for all the remaining loci investigated in the genome wide search. Additional markers (D9S268, D9S162, D9S1684 and D9S942) have been analysed to confirm and better define this positive region. Pairwise linkage data confirmed this locus with a maximum Lod score of 3.67 with marker D9S162 at a recombination frequency of θ=0 (Table 1). Recombinants narrow down a candidate region to approximately 9 cM (as obtained from NCBI database) between marker D9S268 and D9S942. The locus symbol DFNA47 has been assigned to this new NSAD hearing loss locus. The maximal map-specific LOD score obtained with multipoint analysis was 4.26 (Figure 2).
Discussion
In this study we have shown linkage of dominant progressive hearing loss in a large multigeneration Italian kindred coming from Southern Italy to chromosome 9p21–22 (DFNA47). Linkage analysis with highly informative microsatellite markers produced a maximum lod score of 3.67 with marker D9S162 at a recombination frequency of 0. Multipoint linkage analysis further confirms this region with a maximum location score of 4.26. The region defined by recombinants is approximately 9 cM and it contains some known genes. None of them could be considered as a strong candidate but only two weak candidates can be taken into account. The first is SLC24A2 gene, which is a Na-Ca+K-Exchanger, and thus can be hypothetically involved in the homeostasis of endolymphatic fluid while the second one is endophilin 1A gene, which binds dynamin that is expressed in the inner ear. On the contrary, several genes with unknown function are contained in the region and cannot be ruled out as candidate ones. Those functionally related to hearing or expressed in the inner ear are being screened for deafness-causing mutations.
Finally, the identification of a new locus for NSAD hearing loss further confirms the great genetic heterogeneity for this type of deafness making the understanding of molecular bases of hearing loss even more puzzling.
References
Nadol, JB : Hearing loss. N Engl J Med, (1993). 329, 1092–1102.
Davis, AC : Hearing in adults, London: Whurr (1995).
Cohen, Jr MM & Gorlin, RJ : Epidemiology, etiology and genetic patterns. In: Gorlin RJ, Toriello HV, Cohen Jr MM, eds. Hereditary hearing loss and its syndromes, Oxford: Oxford University Press (1995). 9–21.
van Camp, G, Willems, PJ & Smith, RJH : Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet, (1997). 60, 758–764.
Lathrop, GM, Lalouel, JM, Julier, C & Ott, J : Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA, (1984). 81, 3443–3446.
Cottingham, RW, Idury, RM & Shaffer, AA : Faster sequential genetic linkage computations. Am J Hum Genet, (1993). 53, 252–263.
Sobel, E & Lange, K : Descent graphs in pedigree analysis: application to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet, (1996). 58, 1323–1337.
Mukhopadhyay, N, Almsay, L, Schroeder, M, Mulvihill, WP & Weeks, DE : Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet, (1999). 65, abs 436
Acknowledgements
We thank the family for the strong collaboration and help in this work which has been supported in part by Telethon grant E0813 (to P Gasparini), by TIGEM (to P Gasparini) and by IBN-Istituto Banco di Napoli (to P Gasparini).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
D'Adamo, P., Donaudy, F., D'Eustacchio, A. et al. A new locus (DFNA47) for autosomal dominant non-syndromic inherited hearing loss maps to 9p21-22 in a large Italian family. Eur J Hum Genet 11, 121–124 (2003). https://doi.org/10.1038/sj.ejhg.5200929
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200929
Keywords
This article is cited by
-
DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3
European Journal of Human Genetics (2010)
-
Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families
European Journal of Human Genetics (2010)