Abstract
Chromosome 7p15-p14 showed genome-wide significant linkage to asthma related traits among the Finnish and French-Canadian families. As an essential step toward cloning the susceptibility gene, a detailed physical map of the region is needed. In this study we report a dense set of carefully tested, new microsatellite markers for fine mapping embedded in a continuous, easy-to-read, physical map of the region that includes the known genes and putative transcripts. Even though susceptibility genes for asthma are difficult to predict from a multitude of unknown genes mapped to the region, TCRG encoding the γ-chain of the heterodimeric γ/δ T cell receptor is a potential candidate. We present linkage and association results for TCRG in two independent Finnish family sets by using four highly polymorphic microsatellites spanning 169 kb across the locus. Linkage results confirmed our previous findings, but our study did not provide any evidence on behalf of a strong association of TCRG with either high serum total Immunoglobulin (IgE) level or asthma. Our results suggest that some other known or yet unidentified gene in the linkage region is the true asthma susceptibility gene.
Similar content being viewed by others
Introduction
Recent genome wide scans have suggested several regions potentially linked to asthma and related traits. One of the loci of interest is located on 7p15-p14. Among Finnish asthma families, this locus was the only locus that reached genome wide significance level.1 The finding was further confirmed among French-Canadian families and in another independent data set from Finland.1 Daniels et al.2 have reported six regions of possible linkage, including 7p14-p15, and established by simulations that at least some of them are likely to be true positives. German,3 French,4 and Italian families5 have shown some evidence of linkage, but there are also genome scans with inconclusive results,6,7,8 some of which have been done in ethnic populations other than Caucasian.9,10 Furthermore, clinically distinct autoimmune diseases may be controlled by a common set of susceptibility genes.11 7p15-p14 has also been linked to diseases such as multiple sclerosis12 and inflammatory bowel disease,13 and genomic regions homologous to human 7p15-p14 have been linked to insulin dependent diabetes14 and inflammatory arthritis15 in rat models.
Implicating a susceptibility gene for such a complex disorder as asthma is difficult. As an essential step toward identifying a susceptibility gene for asthma, we need a detailed physical map of the linkage region that includes the known genes, putative transcripts, and genetic markers. In this study we report a continuous, easy-to-read, physical map of the region with structural and functional information of known genes. We also present our linkage and association results for TCRG among Finnish asthma families by using four highly polymorphic microsatellite markers within the TCRG locus. Our results suggest that TCRG is not significantly associated with asthma or high serum IgE level, and thus, TCRG might not be the major susceptibility gene for asthma in 7p15-p14.
Materials and methods
Construction of the physical map
We started the construction of a chromosome 7 physical map with the microsatellite markers found to be linked with high serum IgE level and asthma.1 First, we used these markers to determine the corresponding region in the Radiation hybrid (RH) map (http://www.ncbi.nlm.nih.gov/genemap99), and to identify STSs, genes, ESTs, clones and contigs located in the region. Second, we used RH distances of the STS, genes, and ESTs mapped into clones/contigs, and the order of clones/contigs in NBCI Human Chromosome 7 contig map (http://www.ncbi.nlm.nih.gov/genome/seq/chr.cgi?CHR=7) to roughly organise the clones and contigs. Third, to determine the exact order and orientation of clones and contigs, we explored public databases with different contig and clone end sequences, cDNA and EST sequences by BLAST. To fill in the gaps in public sequence, we also used the Celera database (http://public. celera.com/index.cfm). Fourth, we compared the organisation of our map to different genomic databases to make sure that no major discrepancies were present. The comparisons were done using the Human Genome Browser (http://genome.ucsc.edu/goldenPath/octTracks.html), the NCBI Entrez Genomes Database (http://www.ncbi.nlm.nih. gov/genome/seq/), the Sanger Center Ensemble Database (http://www.ensembl.org/), and the Celera database. During the process additional information was also collected from the NCBI databases (http://www.ncbi.nlm.nih.gov/index.html) UniSTS, UniGene, OMIM, LocusLink, and from the Marshfield Genetic Map (http://research.marshfieldclinic. org/genetics). Finally, cDNA sequences were used to map the exons of known genes in genomic sequence. The physical map was drawn based on collected data of clones, genes and markers using the PhysiMap program (http://www.ee. oulu.fi/∼polvi/physimap.htm) developed specifically for this purpose.
Design of new microsatellite markers
To create a dense map of polymorphic markers spread evenly across the linkage region, we screened the publicly available genomic sequence for potentially polymorphic short sequence repeats. The repeats were initially tested by PCR amplification in 10 unrelated individuals. If allelic polymorphism was detected, the Mendelian inheritance was confirmed in 10 nuclear families. Detailed information on primers, PCR conditions and allele frequencies are available in the GenBank (http://www.ncbi.nlm.nih.gov/Genbank/ and at our www site (http://www.genome.helsinki.fi/english/research/projects.xml).
Two independent family collections for linkage and association analysis
First family recruitment was done in the Kainuu province in central eastern Finland in 1994 and 1996. The methods for recruitment, control for population stratification, and clinical evaluation have been described previously in detail.16,17
The second family collection was recruited from the province of North Karelia in southern eastern Finland in 1999. Like Kainuu, it represents the late settlement region of Finland, but with a distinct population history than that of the first data set, ie the settlers came from a different donor population. The study subjects were initially identified by a population survey.1,18 All participants were invited into a clinical trial to evaluate their history of atopic disorders (FinnAllergy study). Probands (n=791) were carefully studied clinically. Among the participants, there were 180 individuals with IgE level >100 kU/l. They were contacted by a letter and asked to join a genetic study with their nuclear family members. Altogether 85 families and 517 individuals were recruited; 26 of the families were trios. In 69 families the proband reported atopic symptoms in multiple family members and other siblings. All participants filled out a questionnaire including questions of respiratory symptoms, smoking habits, medication and physician-made diagnosis of asthma or other atopic disorders.
Total serum IgE level was determined by Diagnostics CAP FEIA (Kabi Pharmacia, Sweden) in both the Kainuu and North Karelia study groups.
Linkage and association analysis for TCRG
TCRG was mapped to the genomic clones AC006033 and AC07245 using exon sequences for V1, V2, V3, V4, V5, V5P, V6, V7, V8, VA, V9, V10, VB, V11, JP1, JP, J1, C1, JP2, J2, and C2 retrieved from the international ImMunoGeneTics database (IMGT) (http://imgt.cines.fr:8104).19 All polymorphic sites (TCR4, TCR6, and TCR7) and two flanking markers D7S2497 (heterozygosity 0.87) and MN35 were genotyped in both sample sets. For genotyping, DNA was extracted from blood leukocytes by a standard non-enzymatic method. The primer pairs were fluorescently labelled (Genosys, UK). PCR amplification was done separately for each marker and run pooled on an ABI377 sequencer. Linkage and association analyses were performed using GENEHUNTER 2.020 and Haplotype Pattern Mining (HPM),21 respectively.
For haplotyping, large pedigrees were divided into trios using an in-house computer programme. The programme identifies the maximal number of trios that are not overlapping and in which one or two members were affected (not both parents). Trios that included members who had not been genotyped or members with unknown phenotype were excluded. Haplotyping was done within each trio and four independent chromosomes were obtained from each trio. In case of ambiguities (missing genotype data, identical heterozygotic genotypes in all of the family members, or Mendel errors), data for that marker were discarded. If the child was affected, the transmitted chromosomes were considered disease associated and the non-transmitted chromosomes as controls. If one of the parents was affected, his/her chromosomes were considered disease associated and the spouse's chromosomes as controls. If both the parent and the child were affected, only the non-transmitted chromosome of an unaffected parent was considered as the control and the other three as disease associated. These haplotypes, labelled as disease-associated or control, were then used as input for HPM.
The HPM algorithm finds all the haplotype patterns up to a given length whose association to the trait, measured by a 2×2 χ2-test, exceeds a given threshold value. To make the algorithm more robust with respect to marker mutations, genotyping errors, and missing data, a user-specified number of gaps is allowed in the patterns. Based on the found patterns, each marker is given a score based on the number of patterns spanning across the marker. HPM estimates the significances of the obtained scores against the null hypothesis that there is no association. The null distributions are estimated by permuting the trait values and recalculating the scores several times. Comparing the observed scores to these distributions yields marker-wise P values that are comparable with each other.
To estimate the overall significance of detected association, accounting simultaneous testing of multiple markers, we performed nested permutation tests. The lowest marker-wise P value is compared to the distribution of the lowest marker-wise P values obtained when the HPM analysis is repeated several times with permuted data, yielding a corrected P value. The procedure has been described in detail (see www.acm.org/sigkdd/kdd2001 and www.cs.helsinki.fi/TR/C.html).22,23
Results
Physical map
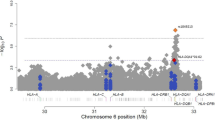
The clones, contigs, genes and markers that are located on 7p15-p14 and linked to asthma are shown in Figure 1. Altogether 184 clones were placed into the map. Nine gaps were detected between the following nine clones:
-
AC006331.2/AC005105.2, AC006027.1/AC005154.1,
-
AC073068.8/AC006033.2, AC073345.10/AC005483.1,
-
AC004987.2/AC072061.8, AC026866.8/AC004988.2,
-
AC011296.2/AC015515.3, AC011738.4/AC005189.1,
-
and AC006454.3/AC004453.1.
The orientation of the following five pairs was determined by using cDNA sequences of genes that had exons in both flanking clones:
-
AC006331.2/AC005105.2, AC006027.1/AC005154.1,
-
AC073345.10/AC005483.1, AC026866.8/AC004988.2,
-
and AC011738.4/AC005189.1.
The genes were Cre-Bpa, NOD1 (OMIM#605980), RPF-1, FLJ11808, and STK17A (OMIM#604726), respectively. The orientation of the remaining four pairs AC073068.8/AC006033.2, AC004987.2/AC072061.8, AC011296.2/AC015515.3 and AC006454.3/AC004453.1, and the length of gaps, was determined using Genomic Scaffold sequences of the public Celera database.
After the assembly of the clones, we compared their order and orientation to the most complete draft assembly of the human genome (http://genome.cse.ucsc.edu/goldenPath/octTracks.html). Draft sequence showed high concordance with our map. However, two major discrepancies were observed. First, NT_000369 showed reverse orientation in the draft sequence, but our assembly results could be confirmed by the Celera and Sanger Institute Ensemble databases. Second, seven clones in our map were absent in the corresponding region of the draft sequence. Five of the clones (AC087072.2, AC087069.2, AC078843.2, AC026866.8, and AC017116.5) could be found in the NCBI Entrez Genome Database placed between the same clones as in our map. In addition, AC015515.3 was shown to partially overlapping with AC027269.5, as in our map. The position of the clone AC087070.2 was confirmed from the Sanger Center Ensemble Database.
When we compared the comprehensive genetic map to our physical map (Figure 2), the order for six markers (D7S1806, D7S1514, D7S668, D7S2469, D7S2548, and D7S678) differed. In 11 cases, two or more markers were mapped into the same genetic position, on our physical map the marker order could be now verified.
Figures 1 and 2 are available in more detail at our web site (http://www.genome.helsinki.fi/eng/research/asthma/pubs.html).
New microsatellite markers
We have carefully tested 53 new microsatellites for the region; 31 of them that were robust to type, polymorphic, and followed Mendelian inheritance in 10 families were chosen for an association study (http://www.genome.helsinki.fi/eng/research/asthma/pubs.html). The intermarker distance for markers shown in the asthma linkage region (Figure 1) is on average 210 kb (range from 17 to 656 kb).
Genes in the region
Altogether 87 known and putative genes were identified in the region (please see http://www.genome.helsinki.fi/eng/research/asthma/pubs.html for specific data). Most genes encode proteins with known structure or/and function, but also genes with only predicted protein product are included. The region includes one cluster of genes (HOXA1-13) belonging to a HOX-gene family while the remaining genes encode proteins that fall functionally and structurally into multiple categories.
TCRG linkage and association analysis
To study the role of TCRG as a potential candidate locus in asthma, we first screened altogether eight tandem repeats within the locus (Figure 3). Three of them (TCR4, TCR6, and TCR7) were confirmed to be highly polymorphic in 20 chromosomes tested and showed Mendelian inheritance in 10 nuclear families. These three markers and two flanking markers D7S2497 and NM35, spanning a 169 kb haplotype, were then genotyped in two Finnish family sets (1150 individuals). Later, however, TCR7 was excluded from the analysis, since sequencing of some individuals with ambiguous genotypes showed that even alleles of same size were actually different in sequence because of several SNPs within the repeat and single nucleotide insertions in a string of adenosines upstream of the repeat. The remaining markers showed strong linkage disequilibrium across the region when analysed in consecutive pairs by 2×2 contingency tables (P<10−28).
Both linkage and haplotype association analyses were done for two phenotypes: asthma and high total serum IgE level, with new markers added. For comparison, exactly the same pedigrees were included into the non-parametric multipoint linkage analysis as in our genome scan.17 For the TCRG markers, the best linkage was found to high IgE level when both data sets were pooled together (NPL score 3.3, P=0.0009, information content 0.86 at D7S2497) (Table 1). For asthma, NPL scores was 2.2 across TCRG (P=0.003, information content 0.80) in the Kainuu data set, among the North Karelia families asthma status was unknown. As expected, the overall linkage for 7p14-p15 was at the same level as before.1 The best linkage hits were outside the TCRG locus; for IgE at D7S691 (NPL score 4.0, P=0.0001, information content 0.80) in the combined data set and for asthma in the Kainuu data set, at GATA31A10 (NPL score 2.7, P=0.004, information content 0.66).
By using the HPM algorithm,21 the best allele and haplotype associations for high IgE were found again in the combined data set. Altogether 596 chromosomes were retrieved from multiplex and nuclear families. Table 2 illustrates the best haplotype associations for high IgE level. All found associations remained rather weak (χ2⩽7.5). For asthma, the number of associated haplotypes and the strength of found associations remained even lower (Table 2).
To study if the overall haplotype distribution in TCRG differs among the disease associated compared to control chromosomes, HPM analysis was done using the following parameters: maximum pattern length four markers, one gap allowed for missing data and possible errors, and χ2 threshold for the association 3.0. The observed scores (range from 6 to 22) (=number of qualified haplotype patterns spanning across the marker) for associated haplotypes remained low for both phenotypes even with this low threshold for association. Permutation test showed no statistically biased haplotype distribution for asthma (marker-wise P values 0.47–0.76) and borderline significance for high IgE level (lowest marker-wise P value 0.038 at TCR6).
To evaluate the overall significance of this smallest marker-wise P value observed, it should be further corrected for multiple testing. Since most of the variables (tested markers and phenotypes) are highly correlated, a simple Bonferroni adjustment is obviously too conservative. To estimate the effect of simultaneous testing of multiple markers, we used a second level permutation test.22,23 The P value of 0.038 was compared to the distribution of the lowest marker-wise P values obtained when the HPM analysis was repeated several times with permuted data, yielding a corrected P value of 0.059. Previously we have estimated by simulations that a correction of 1.3 should be used for the two correlated phenotypes,1 yielding a final P value of 0.08.
Discussion
Chromosome 7p14-p15 has been linked to asthma related traits in Finnish, French-Canadian, and Australian populations1,2 and to other immune mediated disorders.11 For positional cloning projects, an accurate physical map is crucial. Although several maps of the region are publicly available, none of them includes all information and discrepancies exist. Based on our calculations in the Finnish data, the appropriate confidence region for positional cloning of the susceptibility gene for asthma is from the marker D7S526 to D7S1830 (24 cM).24 Our ∼20 Mb physical map covers most of the region with 184 genomic clones and 87 genes. Only information that could be verified from several sources is indicated on the map. Fifty-three new markers were added to 50 previously published markers. They form a highly informative marker map across the region with an average marker density of 210 kb. With a couple of exceptions, physical distances between the markers matched well with the genetic distances (1 cM∼1 Mb), but the more precise map allowed us to order the markers correctly (Figure 2).
As a potentially interesting candidate, TCRG has been mapped to this region.25 TCRG (∼130 kb) consists of a cluster of 14 highly polymorphic variable (V1-V11, V5P, VA and VB, including five pseudogenes), five joining (JP1, JP, J1, JP2, and J2), and two constant genes (C1 and C2) that are rearranged during γ/δ T cell differentiation to encode γ-chain of the heterodimeric T cell receptor19,26 (Figure 3). γ/δ T cells are believed to maintain homeostasis in the airway mucosa during immuno-inflammatory stress and form the first line defence on epithelial surfaces before a α/β T cell response is fully developed.27 Several studies on γ/δ deficient-mice have suggested that γ/δ T cells have an essential role in the development of the IgE response, eosinophilic bronchitis, and bronchial hyperreactivity when mice are sensitised with oval albumine.25,28,29,30 Among asthma patients increased numbers of γ/δ T cells in bronchoalveolar lavage fluid compared to controls have been reported.31,32 γ/δ T cells are potentially also involved in the development of other immune diseases.33,34,35,36,37
Because the germline sequence of the TCRG locus consists of highly homologues elements, genotyping of single nucleotide polymorphisms described in different T cell clones can be extremely difficult.20 We chose to apply in our linkage analysis four highly polymorphic and stable microsatellite markers. Our analysis showed that TCRG is definitively located within the linkage peak that we have reported previously, but the observed NPL scores did not exceed that of the neighbouring markers. Our study did not provide any evidence for a strong association of TCRG with either high serum total IgE level or asthma. All found associations were weak and distributed in multiple, rare TCRG haplotypes. Since asthma is a complex disorder with interactions of many cell types and tissues, it is very difficult to exclude any of the genes of the region. As a new working hypothesis, we must consider the possibility that some other known or yet unidentified genes in the linkage region will show a stronger association to asthma.
References
Laitinen T, Daly MJ, Rioux JD et al. A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population Nat Genet 2001 28: 87–91
Daniels SE, Bhattacharrya S, James A et al. A genome-wide search for quantitative trait loci underlying asthma Nature 1996 383: 247–250
Wjst M, Fischer G, Immervoll T et al. A genome-wide search for linkage to asthma. German Asthma Genetics Group Genomics 1999 58: 1–8
Dizier MH, Besse-Schmittler C, Guilloud-Bataille M et al. Genome screen for asthma and related phenotypes in the French EGEA study Am J Respir Crit Care Med 2000 162: 1812–1818
Malerba G, Venanzi S, Galavotti R et al. Linkage studies to asthma and atopy phenotypes on chromosomes 7, 12, and 19 in the Italian population Am J Hum Genet 2000 67 Suppl 1: 330
Ober C, Tsalenko A, Parry R, Cox NJ . A second-generation genomewide screen for asthma-susceptibility alleles in a founder population Am J Hum Genet 2000 67: 1154–1162
Xu J, Postma DS, Howard TD et al. Major genes regulating total serum immunoglobulin E levels in families with asthma Am J Hum Genet 2000 67: 1163–1173
Mathias RA, Freidhoff LR, Blumenthal MN et al. Genome-wide linkage analyses of total serum IgE using variance components analysis in asthmatic families Genet Epidemiol 2001 20: 340–355
Yokouchi Y, Nukaga Y, Shibasaki M et al. Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31-q33 near the interleukin 12 B locus by a genome-wide search in Japanese families Genomics 2000 66: 152–160
Xu J, Meyers DA, Ober C et al. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma Am J Hum Genet 2001 68: 1437–1446
Becker KG, Simon RM, Bailey-Wilson JE et al. Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases Proc Natl Acad Sci USA 1998 95: 9979–9984
Sawcer S, Jones HB, Feakes R et al. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22 Nat Genet 1996 13: 464–468
Satsangi J, Parkes M, Louis E et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12 Nat Genet 1996 14: 199–202
Jacob HJ, Pettersson A, Wilson D, Mao Y, Lernmark A, Lander ES . Genetic dissection of autoimmune type I diabetes in the BB rat Nat Genet 1992 2: 56–60
Remmers EF, Longman RE, Du Y et al. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats Nat Genet 1996 14: 82–85
Kauppi P, Laitinen LA, Laitinen H, Kere J, Laitinen T . Verification of self-reported asthma and allergy in subjects and their family members volunteering for gene mapping studies Resp Med 1998 92: 1281–1288
Laitinen T, Kauppi P, Ignatius J et al. Genetic control of serum IgE levels and asthma: linkage and linkage disequilibrium studies in an isolated population Hum Mol Genet 1997 6: 2069–2076
Heistaro S, Laatikainen T, Vartiainen E et al. Self-reported health in the Republic of Karelia, Russia and in north Karelia, Finland in 1992 Eur J Public Health 2001 11: 74–80
Lefranc MP . IMGT, the international ImMunoGeneTics database Nucleic Acids Res 2001 29: 207–209
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES . Parametric and nonparametric linkage analysis: a unified multipoint approach Am J Hum Genet 1996 58: 1347–1363
Toivonen HT, Onkamo P, Vasko K et al. Data mining applied to linkage disequilibrium mapping Am J Hum Genet 2000 67: 133–145
Sevon P, Toivonen HTT, Ollikainen V . Tree DT: Gene mapping by tree disequilibrium test in Provost F, Srikant R (eds): KDD-2001 Proceedings of the Seventh ACM SIGKDD International Conference on Knowledge Discovery and Data Mining San Francisco, CA USA: ACM press 2001 pp 365–370
Sevon P, Toivonen HTT, Ollikainen V, Tree DT . Gene mapping by tree disequilibrium test (extended version) Report C 2001 Nr 32:6-7 University of Helsinki, Finland: Department of Computer Science 2001
Kruglyak L, Lander ES . High-resolution genetic mapping of complex traits Am J Hum Genet 1995 56: 1212–1223
Holt PG, Sly PD . Gammadelta T cells provide a breath of fresh air for asthma research Nat Med 1999 5: 1127–1128
Lefranc MP, Rabbitts TH . Genetic organization of the human T cell receptor gamma locus Curr Top Microbiol Immunol 1991 173: 3–9
Kaufmann SH . Gamma/delta and other unconventional T lymphocytes: what do they see and what do they do? Proc Natl Acad Sci USA 1996 93: 2272–2279
Zuany-Amorim C, Ruffie C, Haile S, Vargaftig BB, Pereira P, Pretolani M . Requirement for gammadelta T cells in allergic airway inflammation Science 1998 280: 1265–1267
Lahn M, Kanehiro A, Takeda K et al. Negative regulation of airway responsiveness that is dependent on gammadelta T cells and independent of alphabeta T cells Nat Med 1999 5: 1150–1156
Schramm CM, Puddington L, Yiamouyiannis CA et al. Pro inflammatory roles of T-cell receptor TCRgammadelta and TCRalphabeta lymphocytes in a murine model of asthma Am J Respir Cell Mol Biol 2000 22: 218–225
Spinozzi F, Agea E, Bistoni O et al. Increased allergen-specific, steroid-sensitive gamma delta T cells in bronchoalveolar lavage fluid from patients with asthma Ann Intern Med 1996 124: 223–227
Rossman MD, Carding SR . Gamma delta T cells in asthma Ann Intern Med 1996 124: 266–267
Borsellino G, Koul O, Placido R et al. Evidence for a role of gammadelta T cells in demyelinating diseases as determined by activation states and responses to lipid antigens J Neuroimmunol 2000 107: 124–129
Yeung MM, Melgar S, Baranov V et al. Characterisation of mucosal lymphoid aggregates in ulcerative colitis: immune cell phenotype and TcR-gammadelta expression Gut 2000 47: 215–227
Kanazawa H, Ishiguro Y, Munakata A, Morita T . Multiple accumulation of Vdelta2+ gammadelta T-cell clonotypes in intestinal mucosa from patients with Crohn's disease Dig Dis Sci 2001 46: 410–416
Kretowski A, Mysliwiec J, Kinalska I . Abnormal distribution of gammadelta T lymphocytes in Graves' disease and insulin-dependent diabetes type 1 Arch Immunol Ther Exp 2000 48: 39–42
Olive C, Gatenby PA, Serjeantson SW . Persistence of gamma/delta T cell oligoclonality in the peripheral blood of rheumatoid arthritis patients Immunol Cell Biol 1994 72: 7–11
Acknowledgements
We thank Siv Knaappila, Riitta Lehtinen, and the staff of the Finnish Genome Center for their expertise in genotyping. This study has been supported by the Academy of Finland, the National Technology Agency Tekes, and the Sigrid Jusélius Foundation. J Kere is a member of Biocentrum Helsinki and Center of Excellence for Disease Genetics at University of Helsinki.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Polvi, A., Polvi, T., Sevon, P. et al. Physical map of an asthma susceptibility locus in 7p15-p14 and an association study of TCRG. Eur J Hum Genet 10, 658–665 (2002). https://doi.org/10.1038/sj.ejhg.5200861
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200861