Abstract
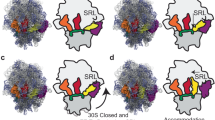
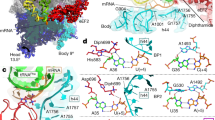
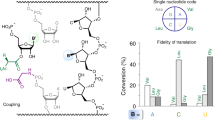
During decoding, a codon of messenger RNA is matched with its cognate aminoacyl-transfer RNA and the amino acid carried by the tRNA is added to the growing protein chain. Here we propose a molecular mechanism for the decoding phase of translation: the transorientation hypothesis. The model incorporates a newly identified tRNA binding site and utilizes a flip between two tRNA anticodon loop structures, the 5′-stacked and the 3′-stacked conformations. The anticodon loop acts as a three-dimensional hinge permitting rotation of the tRNA about a relatively fixed codon–anticodon pair. This rotation, driven by a conformational change in elongation factor Tu involving GTP hydrolysis, transorients the incoming tRNA into the A site from the D site of initial binding and decoding, where it can be proofread and accommodated. The proposed mechanisms are compatible with the known structures, conformations and functions of the ribosome and its component parts including tRNAs and EF-Tu, in both the GTP and GDP states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Garrett, R. A. et al. The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions (ASM, Washington DC, 2000).
Pape, T., Wintermeyer, W. & Rodnina, M. V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 (1998).
Hopfield, J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA 71, 4135–4139 (1974).
Nierhaus, K. H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry 29, 4997–5008 (1990).
Agrawal, R. K. et al. Direct visualization of A-, P-, and E-site transfer RNAs in the E. coli ribosome. Science 271, 1000–1002 (1996).
Wimberly, B. T. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Carter, A. P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000).
Stark, H. et al. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389, 403–406 (1997).
Agrawal, R. K., Penczek, P., Grassucci, R. A. & Frank, J. Visualization of elongation factor G on the Escherichia coli ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA 95, 6134–6138 (1998).
Yusupov, M. M. et al. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 (2001).
Ban, N. et al. the complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Schluenzen, F. et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 (2000).
Nissen, P. et al. Crystal structure of the ternary complex of the Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270, 1464–1472 (1995).
Lake, J. A. Aminoacyl-tRNA binding at the recognition site is the first step of the elongation cycle of protein synthesis. Proc. Natl Acad. Sci. USA 74, 1903–1907 (1977).
Fuller, W. & Hodgson, A. Conformation of the anticodon loop in tRNA. Nature 215, 817–821 (1967).
Woese, C. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature 226, 817–820 (1970).
Crick, F. H. C., Brenner, S., Klug, A. & Pieczenik, G. Speculation on the origin of protein synthesis. Orig. Life 7, 389–397 (1976).
Ladner, J. E. et al. Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl Acad. Sci. USA 72, 4414–4418 (1975).
Quigley, G. J., Seeman, N. C., Wang, A., Suddath, F. L. & Rich, A. Yeast phenylalanine transfer RNA: Atomic coordinates and torsion angles. Nucleic Acids Res. 2, 2329–2349 (1975).
Eiler, S. et al. Synthesis of aspartyl-tRNAAsp in Escherichia coli—a snapshot of the second step. EMBO J. 18, 6532–6541 (1999).
Gupta, S. L., Waterson, J., Sopori, M. L., Weissman, S. M. & Lengyel, P. Movement of the ribosome along the messenger ribonucleic acid during protein synthesis. Biochemistry 10, 4410–4421 (1971).
Thach, S. S. & Thach, R. E. Translocation of messenger RNA and accommodation of fMet-tRNA. Proc. Natl Acad. Sci. USA 68, 1791–1795 (1971).
Harada, F. & Dahlberg, J. E. Specific cleavage of tRNA by nuclease S1. Nucleic Acids Res. 2, 865–871 (1975).
Tal, J. The cleavage of transfer RNA by a single strand specific endonuclease from Neurospora crassa. Nucleic Acids Res. 2, 1073–1082 (1975).
Wrede, P., Woo, N. H. & Rich, A. Initiator tRNAs have a unique anticodon loop conformation. Proc. Natl Acad. Sci. USA 76, 3289–3293 (1979).
Schweisguth, D. C. & Moore, P. B. On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J. Mol. Biol. 267, 505–519 (1997).
Klug, A. et al. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J. Mol. Biol. 131, 669–680 (1979).
Dickerson, R. E. & Chiu, T. K. Helix bending as a factor in protein/DNA recognition. Biopolymers 44, 361–403 (1997).
Carbon, J. & Fleck, E. W. Genetic alteration of structure and function in glycine transfer RNA of Escherichia coli: mechanism of suppression of the tryptophan synthetase A78 mutation. J. Mol. Biol. 85, 371–391 (1974).
Gefter, M. L. & Russell, R. L. Role modifications in tyrosine transfer RNA: a modified base affecting ribosome binding. J. Mol. Biol. 39, 145–157 (1969).
Strycharz, W. A., Nomura, M. & Lake, J. A. Ribosomal proteins L7/L12 localized at a single region of the large subunit by immune electron microscopy. J. Mol. Biol. 126, 123–140 (1978).
Briones, E., Briones, C., Remacha, M. & Ballesta, J. P. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 273, 31956–31961 (1998).
Cundliffe, E. in Structure, Function and Genetics of Ribosomes (eds Hardesty, B. & Kramer, G.) 586–604 (Springer, New York, 1986).
Wimberly, B. T. et al. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell 97, 491–502 (1999).
Markus, M. A. et al. High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA. Nature Struct. Biol. 4, 70–77 (1997).
Stoeffler, G., Cundliffe, E., Stoeffler-Meilicke, M. & Dabbs, E. R. Mutants of Escherichia coli lacking ribosomal protein L11. J. Biol. Chem. 255, 10517–10522 (1980).
Polekhina, G. et al. Helix unwinding in the effector region of elongation factor EF-Tu GDP. Structure 4, 1141–1151 (1996).
Abel, K. et al. An α to β conformational switch in EF-Tu. Structure 4, 1153–1159 (1996).
Song, H. et al. Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol. 285, 1245–1256 (1999).
Abdulkarim, F., Liljas, L. & Hughes, D. Mutations to kirromycin resistance occur in the interface of domains I and III of EF-Tu·GTP. FEBS Lett. 352, 118–122 (1994).
Vogeley, L., Palm, G. J., Mesters, J. R. & Hilgenfeld, R. Conformational change of elongation factor Tu induced by antibiotic binding: crystal structure of the complex between EF-Tu:GDP and aurodox. J. Biol. Chem. 276, 17149–17155 (2001)
Kurland, C. G. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26, 29–50 (1992).
Noller, H. F., Hoffarth, V. & Zimniak, L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419 (1992).
Zaug, A. J. & Cech, T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science 231, 470–475 (1986).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
We thank D. Eisenberg and R. Dickerson for advice and for access to graphics terminals and software, A. Maris and D. Cascio for help, M. Rivera, R. Jain and J. Moore for advice, and M. Kowalczyk for the art work. This work was supported by grants to J.A.L. from the National Institutes of Health, the Astrobiology Institute, the National Science Foundation, and the Department of Energy.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Simonson, A., Lake, J. The transorientation hypothesis for codon recognition during protein synthesis. Nature 416, 281–285 (2002). https://doi.org/10.1038/416281a
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/416281a
This article is cited by
-
Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes
Reviews of Physiology, Biochemistry and Pharmacology (2005)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.