Abstract
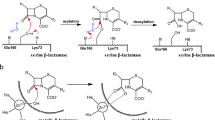
The X-ray crystal structure of the molecular complex of penicillin G with a deacylation-defective mutant of the RTEM-1 β-lactamase from Escherichia coli shows how these antibiotics are recognized and destroyed. Penicillin G is covalently bound to Ser 70 Oγ as an acyl-enzyme intermediate. The deduced catalytic mechanism uses Ser 70 Oγ as the attacking nucleophile during acylation. Lys 73 Nζ acts as a general base in abstracting a proton from Ser 70 and transferring it to the thiazolidine ring nitrogen atom via Ser 130 Oγ. Deacylation is accomplished by nucleophilic attack on the penicilloyl carbonyl carbon by a water molecule assisted by the general base, Glu 166.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Herzberg, O. & Moult, J. Curr. Opin. struct. Biol. 1, 946–953 (1991).
Frère, J.-M. & Joris, B. Crit. Rev. Microbiol. 11, 299–396 (1985).
Ghuysen, J.-M. A. Rev. Microbiol. 45, 37–67 (1991).
Herzberg, O. & Moult, J. Science 236, 694–701 (1987).
Herzberg, O. J. molec. Biol. 217, 701–719 (1991).
Moews, P. C., Knox, J. R., Dideberg, O., Charlier, P. & Frère, J.-M., Proteins Struct. Funct Genet. 7, 156–171 (1990).
Knox, J. R. & Moews, P. C. J. molec. Biol. 220, 435–455 (1991).
Jelsch, C., Lenfant, F., Masson, J. M. & Samama, J. P. FEBS Lett 299, 135–142 (1992).
Knowles, J. R. Acct. Chem. Res. 18, 97–104 (1985).
Christensen, H., Martin, M. T. & Waley, S. G. Biochem. J. 266, 853–861 (1990).
Gibson, R. M., Christensen, H. & Waley, S. G. Biochem. J. 272, 613–619 (1990).
Adachi, H., Ohta, T. & Matsuzawa, H. J. biol. Chem. 266, 3186–3191 (1991).
Escobar, W. A., Tan, A. K. & Fink, A. L. Biochemistry 30, 10783–10787 (1991).
Ellerby, L. M., Escobar, W. A., Fink, A. L., Mitchinson, C. & Wells, J. A. Biochemistry 29, 5797–5800 (1990).
Dalbadie-McFarland, G., Neitzel, J. J. & Richards, J. H. Biochemistry 25, 332–338 (1986).
Martin, M. T. & Waley, S. G. Biochem. J. 254, 923–925 (1988).
Virden, R., Tan, A. K. & Fink, A. L. Biochemistry 29, 145–153 (1990).
Healey, W. J., Labgold, M. R. & Richards, J. H. Proteins Struct. Funct. Genet. 6, 275–283 (1989).
Fisher, J., Belsaco, J. G., Khosla, S. & Knowles, J. R. Biochemistry 19, 2895–2901 (1980).
Jacob, F. et al. Protein Engng 4, 79–86 (1990).
Jacob, F., Joris, B., Lepage, S., Dusart, J. & Frére, J.-M. Biochem. J. 271, 399–406 (1990).
Cartwright, S. J., Tan, A. K. & Fink, A. L. Biochem. J. 263, 905–912 (1989).
Abraham, E. P. & Chain, E. Nature 146, 837 (1940).
Matthew, M. & Hedges, R. W. J. Bact. 125, 713–718 (1976).
Green, D. W., Ingram, V. M. & Perutz, M. F. Proc. R. Soc. A225, 287–307 (1954).
Rossman, M. G. The Molecular Replacement Method. Int. Sci. Rev. Vol. 13 (Gordon & Breach, New York, 1972).
Bernstein, F. C. et al. J. molec. Biol. 112, 535–542 (1977).
Ambler, R. P. et al. Biochem. J. 276, 269–272 (1991).
Henderson, R. J. molec. Biol. 54, 341–354 (1970).
Robertus, J. D., Kraut, J., Alden, R. A. & Birktoft, J. J. Biochemistry 11, 4293–4303 (1972).
Crowfoot, D., Bunn, C. W., Rogers-Low, B. W. & Turner-Jones, A. in The Chemistry of Penicillin (eds Clarke, H. T., Johnson, J. R. & Robinson, R.) 310–367 (Princeton Univ, Press, 1949).
Dexter, D. D. & van der Veen, J. M. J. chem. Soc. 185, 110–115 (1978).
Oefner, G. et al. Nature 343, 284–288 (1990).
Zafaralla, G., Manavathu, E. K., Lerner, S. A. & Mobashery, S. Biochemistry 31, 3847–3852 (1992).
Kraut, J. A. Rev. Biochem. 46, 331–358 (1977).
Knap, A. K. & Pratt, R. F. Biochem. J. 273, 85–91 (1991).
James, M. N. G., Hall, D. & Hodgkin, D. C. Nature 220, 168–170 (1968).
Sweet, R. M. & Dahl, L. F. J. Am. chem. Soc. 92, 5489–5507 (1970).
Viera, J. & Messing, J. Meth. Enzym 153, 3–11 (1987).
Hamlin, R. Meth. Enzym. 114, 416–452 (1985).
Howard, A. J., Nielson, C. & Xuong, N. H. Meth. Enzym. 114, 452–472 (1985).
Otwinowski, Z. Am. Crystallogr. Ass. Ann. Meeting, New Orleans, Abstr. CO4 (1990).
Navaza, J. Acta crystallogr. A46, 619–620 (1990).
Read, R. J. & Schierbeek, A. J. J. appl. Crystallogr. 21, 490–495 (1988).
Read, R. J. Acta crystallogr. A42, 140–149 (1986).
Jones, T. A. J. Appl. Crystallogr. 11, 268–272 (1978).
Leslie, A. G. W., Brick, P. & Wonacott, A. J. CCP4 Newsletter 18, 33–39 (1986).
Brünger, A. J. J. molec. Biol. 203, 803–816 (1988).
Hendrickson, W. A. & Konnert, J. H. in Computing in Crystallography (eds Diamond, R., Ramaseshan, S. & Venkatesan, K.) 1301–1323 (Indian Institute of Sciences, Bangalore 1980).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Strynadka, N., Adachi, H., Jensen, S. et al. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature 359, 700–705 (1992). https://doi.org/10.1038/359700a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/359700a0
This article is cited by
-
Mapping the determinants of catalysis and substrate specificity of the antibiotic resistance enzyme CTX-M β-lactamase
Communications Biology (2023)
-
FPocketWeb: protein pocket hunting in a web browser
Journal of Cheminformatics (2022)
-
Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as β-lactamase inhibitors
Scientific Reports (2022)
-
An in vivo selection system with tightly regulated gene expression enables directed evolution of highly efficient enzymes
Scientific Reports (2021)
-
Unraveling the energetic significance of chemical events in enzyme catalysis via machine-learning based regression approach
Communications Chemistry (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.