Abstract
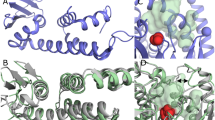
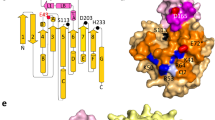
INTERFACIAL adsorption of pancreatic lipase is strongly dependent on the physical chemical properties of the lipid surface. These properties are affected by amphiphiles such as phospholipids and bile salts. In the presence of such amphiphiles, lipase binding to the interface requires a protein cofactor, colipase1–3. We obtained crystals of the pancreatic lipase–procolipase complex and solved the structure at 3.04 Å resolution. Here we describe the structure of procolipase, which essentially consists of three 'fingers' and is topologically comparable to snake toxins4,5. The tips of the fingers contain most of the hydrophobic amino acids and presumably form the interfacial binding site. Lipase binding occurs at the opposite side to this site and involves polar interactions. Determination of the three-dimensional structure of pancreatic lipase has revealed the presence of two domains: an amino-terminal domain, at residues 1–336 containing the active site and a carboxy-terminal domain at residues 337–449 (ref. 6). Procolipase binds exclusively to the C-terminal domain of lipase. No conformational change in the lipase molecule is induced by the binding of procolipase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Verger, R. Lipases 83–150 (eds Borgström, B. & Brockman, H.) (Elsevier, Amsterdam, 1984).
Borgström, B. & Erlanson-Albertsson, C. in Lipases (eds Borgström, B. & Brockman, H.) 151–183 (Elsevier, Amsterdam, 1984).
Sarda, L. in Lipases: Structure, Mechanism and Genetic Engineering Vol. 16 (eds Alberghima et al) 145–155 (GBF monographs, VCH, Weinheim, 1991).
Rees, B. et al. Proc. natn Acad. Sci. USA. 84, 3132–3136 (1987).
Bourne, P. E. et al. Eur. J. Biochem. 153, 521–527 (1985).
Winkler, F. K., D'Arcy, A. & Hunziker, W. Nature 343, 771–774 (1990).
Rathelot, J. et al. Biochimie 63, 227–234 (1981).
Erlanson, C., Charles, M., Astier, M. & Desnuelle, P. Biochim. blophys. Acta 259, 198–203 (1974).
Donner, J., Spink, C., Borgström, B. & Sjöholm, I. Biochemistry 15, 5413–5417 (1976).
Kabsch, W. & Sander, C. Biopolymers 22, 2577–2637 (1983).
Janin, J. & Chothia, C. J. biol. Chem. 256, 16027–16030 (1990).
Abousalham, A., Chaillan, C., Kerfelec, B., Foglizzo, E. & Chapus, C. Protein Engng. 5, 105–111 (1992).
Cozzone, P. J., Canioni, P., Sarda, L. & Kaptein, R. Eur. J. Biochem. 114, 119–126 (1984).
Mclntyre, J. C., Hundley, P. & Behnke, W. D. Biochem. J. 245, 821–829 (1987).
Brzozowski, A. M. et al. Nature 351, 491–494 (1991).
Derewenda, U., Brzozowski, A. M., Lawson, D. M. & Derewenda, Z. S. Biochemistry 31, 1532–1541 (1992).
Brünger, A. T. Acta crystallogr. 46, 46–57 (1990).
Roussel, A. & Cambillau, C. in Silicon Graphics Directory (Silicon Graphics, Mountain View, California, 1991).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
van Tilbeurgh, H., Sarda, L., Verger, R. et al. Structure of the pancreatic lipase–procolipase complex. Nature 359, 159–162 (1992). https://doi.org/10.1038/359159a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/359159a0
This article is cited by
-
Structure of dimeric lipoprotein lipase reveals a pore adjacent to the active site
Nature Communications (2023)
-
Exploring inhibition mechanism and nature of lipase by Ligupurpuroside A extracted from Ku-Ding tea
Medicinal Chemistry Research (2018)
-
Inhibition of pancreatic lipase by berberine and dihydroberberine: an investigation by docking simulation and experimental validation
Medicinal Chemistry Research (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.