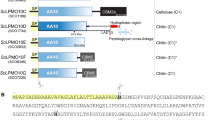
Abstract
CELLULOSIC biomass is recycled by a variety of microorganisms occupying different habitats1. Studies of their cellulase systems have included the purification of enzyme components, the determination of their enzymological properties2 and the cloning and characterization of their structural genes3. Sequence analysis of more than 70 cellulases permits grouping into seven families corresponding to distinct structural types4,5. The three-dimensional structure of the catalytic core of cellobiohydrolase CBHII from the fungus Trichoderma reesei has been reported6. Here we show that endoglucanase CelD from Clostridium thermocellum, which is representative of a different family of cellulose-degrading enzymes consisting of at least 11 bacterial, fungal and plant endoglucanases5,7, has a globular structure, with an amino-terminal immunoglobulin-like domain tightly packed against a larger catalytic domain. The latter shows a novel protein fold, shaped like an α-barrel of 12 helices connected by loops that form the active site. The structure of a complex CelD with a substrate analogue suggests a mechanism for substrate hydrolysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Ljungdahl, L. G. & Eriksson, K. E. Adv. Microbiol. Ecol. 8, 237–299 (1985).
Coughlan, M. P. Biotechnol. Genet. Engng Rev. 3, 39–109 (1985).
Béguin, P. A. Rev. Microbiol. 44, 219–248 (1990).
Henrissat, B., Claeyssens, M., Tomme, P., Lemesle, L. & Mornon, J.-P. Gene 81, 83–95 (1989).
Gilkes, N. R., Henrissat, B., Kilburn, D. G., Miller, R. C. & Warren, R. A. J. Microbiol. Rev. 55, 303–315 (1991).
Rouvinen, J., Bergfors, T., Teeri, T., Knowles, J. K. C. & Jones, T. A. Science 249, 380–386 (1990).
Navarro, A., Chebrou, M.-C., Béguin, P. & Aubert, J.-P. Res. Microbiol. 142, 927–936 (1991).
Joliff, G. et al. Biotechnology 4, 896–900 (1986).
Tokatlidis, K., Salamitou, S., Beguin, P., Dhurjati, P. & Aubert, J.-P. FEBS Lett. 291, 185–188 (1991).
Kretsinger, R. H. Cold Spring Harb. Symp. quant. Biol. 52, 499–510 (1987).
Lascombe, M.-B. et al. Proc. natn. Acad. Sci. U.S.A. 86, 607–611 (1989).
Deisenhofer, J. Biochemistry 20, 2361–2370 (1981).
Tomme, P. thesis, Univ. Ghent (1991).
Vyas, N. K. Curr. Opin. struct. Biol. 1, 732–740 (1991).
Tomme, P. et al. J. biol. Chem. 266, 10313–10318 (1991).
Chauvaux, S., Béguin, P. & Aubert, J.-P. J. biol. Chem. (in the press).
Kelly, J. A., Sielecki, A. R., Sykes, B. D., James, M. N. G. & Phillips, D. C. Nature 282, 875–878 (1979).
Gebbler, J. et al. J. biol. Chem. (in the press).
Joliff, G., Béguin, P. & Aubert, J.-P. Nucleic Acids Res. 14, 8605–8613 (1986).
Joliff, G. et al. J. molec. Biol. 189, 249–250 (1986).
Kabsch, W. J. appl. Crystallogr. 21, 916–924 (1988).
CCP4 The S.E.R.C. Collaborative Computing Project No. 4 (Daresbury Laboratories, Warrington, UK, 1979).
Jones, T. A. J. appl. Crystallogr. 11, 268–272 (1978).
Greer, J. Meth. Enzym. 115, 206–224 (1985).
Jones, T. A. & Thirup, S. EMBO J. 5, 819–822 (1986).
Brünger, A. T. J. molec. Biol. 203, 803–816 (1988).
Priestle, J. P. J. appl. Crystallogr. 21, 572–576 (1988).
Chothia, C. & Lesk, A. M. EMBO J. 5, 823–826 (1986).
Holmgren, A. & Bränden, C.-I. Nature 342, 248–251 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Juy, M., Amrt, A., Alzari, P. et al. Three-dimensional structure of a thermostable bacterial cellulase. Nature 357, 89–91 (1992). https://doi.org/10.1038/357089a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/357089a0
This article is cited by
-
New thermostable endoglucanase from Spirochaeta thermophila and its mutants with altered substrate preferences
Applied Microbiology and Biotechnology (2021)
-
Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action
Nature Communications (2019)
-
Characterization of a Theme C Glycoside Hydrolase Family 9 Endo-Beta-Glucanase from a Biogas Reactor Metagenome
The Protein Journal (2018)
-
Comparative characterization of all cellulosomal cellulases from Clostridium thermocellum reveals high diversity in endoglucanase product formation essential for complex activity
Biotechnology for Biofuels (2017)
-
A Novel Endoglucanase (Cel9P) From a Marine Bacterium Paenibacillus sp. BME-14
Applied Biochemistry and Biotechnology (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.