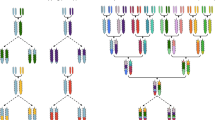
Key Points
-
Geneticists are poised to move into a dissection of complex phenotypic traits.
-
The complexity of phenotypes probably arises from the segregation of alleles at many, interacting loci (quantitative trait loci, or QTL), the effects of which are sensitive to the environment.
-
QTL mapping will allow biologists to make the first steps towards understanding the genetics and evolution of complex phenotypic traits.
-
At its most basic level, QTL mapping simply involves finding an association between a genetic marker and a phenotype that can be measured.
-
Generally, QTL mapping requires two parental strains with different alleles that affect variation in a single trait and a polymorphic genetic map that allows the two strains to be distinguished genetically.
-
QTL analysis of adaptive traits has revealed the number of genes that underlie those traits, their mode of action, and their dependence on the starting material and the environment.
-
QTL analysis has provided vital insights into the processes of speciation and plant domestication, as well as having enormous practical potential for agriculture.
-
The fw2.2 QTL for fruit size in tomato has been characterized at the molecular level.
-
Comparative QTL mapping has revealed insights into plant domestication and has also shown how the merger of genomes of divergent evolutionary histories can produce 'unique avenues' for selection.
-
There are numerous caveats for the application of QTL mapping techniques, including the expense of map construction, the bias in location and effect of individual QTL, and the difficulty in moving from QTL to an actual genetic locus.
-
QTL analysis is as applicable to humans and animals as it is to plants, although working in a plant system has many advantages, including the ability to generate large numbers of progeny from inbred parental lines and to work in ecologically relevant environments.
Abstract
Gregor Mendel was either clever or lucky enough to study traits of simple inheritance in his pea plants; however, many plant characters of interest to modern geneticists are decidedly complex. Understanding the genetic basis of such complex, or quantitative, traits requires a combination of modern molecular genetic techniques and powerful statistical methods. These approaches have begun to give us insight into understanding the evolution of complex traits both in crops and in wild plants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kaul, S. et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796– 815 (2000).
Theologis, A. et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816– 820 (2000).
Lin, X. Y. et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761– 768 (1999).
Salanoubat, M. et al. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408, 820– 822 (2000).
Mayer, K. et al. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769– 777 (1999).
Tabata, S. et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823– 826 (2000).
Adam, D. Now for the hard ones. Nature 408, 792– 793 (2000).
Bulmer, M. G. The Mathematical Theory of Quantitative Genetics (Clarendon, Oxford, 1985).
Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics (Addison–Wesley–Longman, Harlow, 1996).
Lynch, M. & Walsh, B. Genetics and the Analysis of Quantitative Traits (Sinauer Associates, Sunderland, Massachusetts, 1997).
Tanksley, S. D. Mapping polygenes. Annu. Rev. Genet. 27, 205–233 (1993).
Liu, B.-H. Statistical Genomics: Linkage, Mapping and QTL Analysis (Boca Raton, Florida, USA, 1998).An excellent overview of QTL mapping techniques.
Burr, B. & Burr, F. A. Recombinant inbreds for molecular mapping in maize: theoretical and practical considerations. Trends Genet. 7, 55–60 ( 1991).
Moreno-Gonzalez, J. Efficiency of generations for estimating marker-associated QTL effects by multiple regression. Genetics 135, 223– 231 (1993).
Sax, K. The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8, 552– 560 (1923).The first mapping of a QTL by association with a Mendelian marker.
Lister, C. & Dean, C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4, 745–750 ( 1993).
Cho, R. J. et al. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nature Genet. 23, 203– 207 (1999).
Barton, N. H. & Turelli, M. Evolutionary quantitative genetics: how little do we know? Annu. Rev. Genet. 23, 337–370 (1989).
Lande, R. The response to selection on major and minor mutations affecting a metrical trait. Heredity 50, 47– 65 (1983).
Orr, H. A. & Coyne, J. A. The genetics of adaptation: a reassessment . Am. Nat. 140, 725–742 (1992).The landmark paper that exhumed the argument that genes of major effect could be important in adaptation.
Fisher, R. A. The Genetical Theory of Natural Selection (Dover, New York, 1958).
Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C. J. & Koornneef, M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 96, 4710–4717 ( 1999).
Stanton, M. L. Seed variation in wild radish: effect of seed size on components of seedling and adult fitness. Ecology 65, 1105– 1112 (1984).
Fisher, R. A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 7, 179–188 ( 1936).
Juenger, T., Purugganan, M. D. & Mackay, T. F. C. Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics 156, 1379–1392 (2000).
Kelly, M. G. & Levin, D. A. Directional selection on initial flowering date in Phlox drummondii (Polemoniaceae). Am. J. Bot. 87, 382–391 ( 2000).
Kowalski, S. P., Lan, T. H., Feldmann, K. A. & Paterson, A. H. QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol. Gen. Genet. 245, 548– 555 (1994).
Mitchell-Olds, T. Genetic constraints on life history evolution — quantitative trait loci influencing growth and flowering in Arabidopsis thaliana. Evolution 50, 140–145 ( 1996).
Kuittinen, H., Sillanpää, M. J. & Savolainen, O. Genetic basis of adaptation: flowering time in Arabidopsis thaliana. Theor. Appl. Genet. 95, 573–583 (1997).
Byrne, M. et al. Identification and mode of action of quantitative trait loci affecting seedling height and leaf area in Eucalyptus nitens. Theor. Appl. Genet. 94, 674–681 (1997).
Byrne, M., Murrell, J. C., Owen, J. V., Williams, E. R. & Moran, G. F. Mapping of quantitative trait loci influencing frost tolerance in Eucalyptus nitens. Theor. Appl. Genet. 95, 975–979 (1997).
Hurme, P., Silanpää, M. J., Arjas, E., Repo, T. & Savolainen, O. Genetic basis of climatic adaptation in Scots pine by Bayesian quantitative trait locus analysis. Genetics 156, 1309–1322 ( 2000).
Bradshaw, H. D. Jr & Stettler, R. Molecular genetics of growth and development in Populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics 139, 963– 973 (1995).
Frewen, B. E. et al. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154, 837–845 (2000).
Lewontin, R. C. The Genetic Basis of Evolutionary Change (Columbia University Press, New York, 1974).
Gottlieb, L. D. Genetics and morphological evolution in plants. Am. Nat. 123, 681–709 (1984). An early, but underappreciated, paper that raised the issue that genes of major effect could be important in speciation.
Coyne, J. A. & Lande, R. The genetic basis of species differences in plants. Am. Nat. 126, 141– 145 (1985).
Bradshaw, H. D. Jr, Wilbert, S. M., Otto, K. G. & Schemske, D. W. Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus). Nature 376, 762–765 ( 1995).One of the first attempts in natural populations to identify genes that are important in speciation by QTL mapping.
Bradshaw, H. D. Jr, Otto, K. G., Frewen, B. E., McKay, J. K. & Schemske, D. W. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149, 367–382 (1998).
Schemske, D. W. & Bradshaw, H. D. Jr. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proc. Natl Acad. Sci. USA 96, 11910–11915 (1999).
Kelly, A. J. & Willis, J. H. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol. Ecol. 7, 769–774 (1998).
Cruzan, M. B. & Arnold, M. L. Ecological and genetic associations in an Iris hybrid zone. Evolution 47, 1432–1445 (1993).
Arnold, M. L. Anderson's paradigm: Louisiana irises and the study of evolutionary phenomena . Mol. Ecol. 9, 1687–1698 (2000).
Hodges, S. A. & Arnold, M. L. Floral and ecological isolation between Aquilegia formosa and Aquilegia pubescens. Proc. Natl Acad. Sci. USA 91, 2493– 2496 (1994).
Rieseberg, L. H., Sinervo, B., Linder, C. R., Ungerer, M. C. & Arias, D. M. Role of gene interactions in hydrid speciation: evidence from ancient and experimental hybrids. Science 272, 741–745 ( 1996).
Kim, S.-C. & Rieseberg, L. H. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression . Genetics 153, 965–977 (1999).
Rieseberg, L. H., Whitton, J. & Gardner, K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152, 713–977 (1999).
Hodges, S. A., Burke, J. M. & Arnold, M. L. Natural formation of Iris hybrids: experimental evidence on the establishment of hybrid zones. Evolution 50, 2504–2509 (1996).
Stuber, C. W. Mapping and manipulating quantitative traits in maize. Trends Genet. 11, 477–481 ( 1995).
Young, N. D. A cautiously optimistic vision for marker-assisted breeding. Mol. Breed. 5, 505–510 ( 1999).
Grandillo, S., Ku, H. M. & Tanksley, S. D. Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 99, 978–987 (1999).
Alpert, K. B., Grandillo, S. & Tanksley, S. D. fw-2.2 — a major QTL controlling fruit weight is common to both red-fruited and green-fruited tomato species. Theor. Appl. Genet. 91, 994–1000 (1995).
Alpert, K. B. & Tanksley, S. D. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: a major fruit weight quantitative trait locus in tomato. Proc. Natl Acad. Sci. USA 93, 15503–15507 (1996).
Frary, A. et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85– 88 (2000).A landmark in QTL analysis: the first molecular characterization of a locus that was originally identified entirely by QTL mapping. References 51 – 53 are the prologue to this milestone.
Doebley, J. & Stec, A. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129 , 285–295 (1991).
Doebley, J. Mapping the genes that made maize. Trends Genet. 8, 302–307 (1992).
Doebley, J. & Stec, A. Inheritance of the morphological differences between maize and teosinte: comparison of results for two F2 populations . Genetics 134, 559–570 (1993).
Doebley, J., Stec, A. & Gustus, C. Teosinte branched 1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333–346 (1995).
White, S. & Doebley, J. Of genes and genomes and the origin of maize. Trends Genet. 14, 327– 332 (1998).
Dorweiler, J., Stec, A., Kermicle, J. & Doebley, J. Teosinte glume architecture 1: a genetic locus controlling a key step in maize evolution . Science 262, 233–235 (1993).
Doebley, J., Stec, A. & Hubbard, L. The evolution of apical dominance in maize. Nature 386, 485–488 ( 1997).
Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. The limits of selection during maize domestication. Nature 398, 236–239 ( 1999).References 55 – 62 chronicle a decade of work on understanding the genetics of the domestication of modern maize.
Paterson, A. H. et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269, 1714–1718 (1995). An excellent example of the power of comparative QTL mapping.
Ramsey, J. & Schemske, D. W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501 ( 1998).
Soltis, P. S. & Soltis, D. E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl Acad. Sci. USA 97, 7051–7057 ( 2000).
Hilu, K. W. Polyploidy and the evolution of domesticated plants. Am. J. Bot. 80, 1494–1499 ( 1993).
Paterson, A. H. et al. Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1539 ( 2000).
Jiang, C. X., Wright, R. J., El-Zik, K. M. & Paterson, A. H. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl Acad. Sci. USA 95 , 4419–4424 (1998).
Wright, R. J., Thaxton, P. M., El-Zik, K. M. & Paterson, A. H. D-subgenome bias of Xcm resistance genes in tetraploid Gossypium (cotton) suggests that polyploid formation has created novel avenues for evolution. Genetics 149, 1987– 1996 (1998).
Beavis, W. D. The power and deceit of QTL experiments: lessons from comparative QTL studies . Proc. Corn and Sorghum Industry Res. Conf., Am. Seed Trade Assoc., Washington DC, 255–266 ( 1994).
Beavis, W. D. in Molecular Dissection of Complex Traits (ed. Paterson, A. H.) 145 –162 (CRC, Boca Raton, Florida, 1998 ).References 70 and 71 provided the first and most influential caveats for the use of QTL analysis in both agriculture and evolutionary biology.
Copenhaver, G. P., Browne, W. E. & Preuss, D. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl Acad. Sci. USA 95, 247–252 (1998).
Begun, D. J. & Aquadro, C. F. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster . Nature 356, 519–520 (1992).
Risch, N. J. Searching for genetic determinants in the new millennium. Nature 405, 847–856 ( 2000).
Weiss, K. M. & Terwilliger, J. D. How many diseases does it take to map a gene with SNPs? Nature Genet. 26, 151–157 (2000).
Cardon, L. R. & Bell, J. I. Association study designs for complex diseases. Nature Rev. Genet. 2, 91– 99 (2001).
Doerge, R. W., Zeng, Z. B. & Weir, B. S. Statistical issues in the search for genes affecting quantitative traits in experimental populations. Stat. Sci. 12, 195–219 (1997). An excellent overview of the statistical issues involved in QTL analysis.
Melchinger, A. E., Utz, H. F. & Schon, C. C. Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149, 383–403 ( 1998).
Otto, S. P. & Jones, C. D. Detecting the undetected: estimating the total number of loci underlying a quantitative trait. Genetics 156, 2093–2107 ( 2000).
Paterson, A. H. et al. Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127, 181–197 ( 1991).
Jansen, R. C. Interval mapping of multiple quantitative trait loci. Genetics 135, 205–211 ( 1993).
Lander, E. & Kruglyak, L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genet. 11, 241–247 ( 1995).
Lander, E. S. & Botstein, D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121, 185–199 (1989). In this paper, interval mapping with molecular markers to map QTL was first proposed for species in which many morphological markers were unavailable. A maximum-likelihood statistical approach for QTL mapping was also developed.
Knott, S. A. & Haley, C. S. Maximum likelihood mapping of quantitative trait loci using full-sib families. Genetics 132, 1211–1222 (1992).
Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 136, 1457–1468 (1994).
Churchill, G. A. & Doerge, R. W. Empirical threshold values for quantitative trait mapping. Genetics 138 , 963–971 (1994).
Doerge, R. W. & Churchill, G. A. Permutation tests for multiple loci affecting a quantitative character. Genetics 142 , 285–294 (1996).
Sillanpää, M. J. & Arjas, E. Bayesian mapping of multiple quantitative trait loci from incomplete inbred line cross data . Genetics 148, 1373–1388 (1998).
Haley, C. S., Knott, S. A. & Elsen, J.-M. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics 136, 1195–1207 (1994).
Hoeschele, I., Uimari, P., Grignola, F. E., Zhang, Q. & Gage, K. M. Advances in statistical methods to map quantitative trait loci in outbred populations. Genetics 147, 1445–1457 ( 1997).
Sillanpää, M. J. & Arjas, E. Bayesian mapping of multiple quantitative trait loci from incomplete outbred offspring data . Genetics 151, 1605–1619 (1999).
Pérez-Enciso, M. & Varona, L. Quantitative trait loci mapping in F2 crosses between outbred lines. Genetics 155, 391–405 ( 2000).
George, A. W., Visscher, P. M. & Haley, C. S. Mapping quantitative trait loci in complex pedigrees: a two-step variance component approach. Genetics 156 , 2081–2092 (2000).
Hoeschele, I. in Handbook of Statistical Genetics (eds Balding, D., Bishop, M. & Cannings, C.) 599–644 (John Wiley & Sons Ltd, London, 2001).
Liu, S.-C., Lin, Y.-R., Irvine, J. E. & Paterson, A. H. in Molecular Dissection of Complex Traits (ed. Paterson, A. H.) 95–101 (CRC, Boca Raton, Florida, 1998).
Ming, R. et al. Detailed alignment of Saccharum and Sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes . Genetics 150, 1663–1682 (1998).
Doerge, R. W. & Craig, B. A. Model selection for quantitative trait locus analysis in polyploids. Proc. Natl Acad. Sci. USA 97, 7951–7956 (2000).
Rieseberg, L. H. & Buerkle, C. A. Genetic mapping in hybrid zones. Am. Nat. (in the press).
Provine, W. B. The Origins of Theoretical Population Genetics (Chicago Univ. Press, Illinois, 1971).A superb history of the early conflict between the 'Mendelians' and the 'biometricians', and its resolution.
East, E. M. A Mendelian interpretation of variation that is apparently continuous. Am. Nat. 44, 65–82 ( 1910).
Fisher, R. A. The correlation between relatives on the supposition of Mendelian inheritance . Trans. R. Soc. Edinb. 52, 399– 433 (1918).
Lande, R. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 26, 221– 235 (1976).
Lande, R. Quantitative genetic analysis of multivariate evolution, applied to brain:body size allometry. Evolution 33, 402– 416 (1979).
Lande, R. & Arnold, S. J. The measurement of selection on correlated characters. Evolution 37, 1210 –1226 (1983).
Via, S. & Lande, R. Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522 (1985).
Turelli, M. & Barton, N. H. Dynamics of polygenic characters under selection. Theor. Popul. Biol. 38, 1–57 (1990).
Barton, N. H. & Turelli, M. Natural and sexual selection on many loci. Genetics 127, 229 –255 (1991).
Turelli, M. & Barton, N. H. Genetic and statistical analyses of strong selection on polygenic traits: what, me normal? Genetics 138, 913–941 ( 1994).
Robertson, A. in Heritage from Mendel (ed. Brink, A.) 265– 280 (Wisconsin Univ. Press, Madison, Wisconsin, 1967 ).
Haley, C. S. & Knott, S. A. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69, 315–324 ( 1992).
Thoday, J. M. Location of polygenes. Nature 191, 368– 370 (1961).
Zeng, Z.-B. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl Acad. Sci. USA 90, 10972–10976 (1993).
Kao, C. H., Zeng, Z.-B. & Teasdale, R. D. Multiple interval mapping for quantitative trait loci. Genetics 152, 1203– 1216 (1999).
Zeng, Z.-B., Kao, C. H. & Basten, C. J. Estimating the genetic architecture of quantitative traits. Genet. Res. 74, 279– 289 (1999).
Basten, C. J., Weir, B. S. & Zeng, Z.-B. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping (Department of Statistics, North Carolina Univ. Press, Raleigh, North Carolina, 1995).
Reinisch, A. J. et al. A detailed RFLP map of cotton, Gossypium hirsutum x Gossypium barbadense: chromosome organization and evolution in a disomic polyploid genome. Genetics 138, 829– 847 (1994).
Acknowledgements
I thank M. Arnold, R. Baucom, A. Bouck, C. Goodwillie, A. Johnson, A. Paterson, L. Rieseberg and J. Willis for unpublished material, helpful discussions and constructive comments on the manuscript.
Author information
Authors and Affiliations
Related links
Related links
FURTHER INFORMATION
Arabidopsis Biological Resource Centre (ABRC)
Cereon Genomics Arabidopsis SNP collection
Nottingham Arabidopsis Stock Centre: Columbia × Landsberg RI lines
Rockefeller University's collection of genetic analysis software
The Arabidopsis Information Resource (TAIR)
The Institute for Genomic Research
ENCYCLOPEDIA OF LIFE SCIENCES
Glossary
- MULTIVARIATE NORMAL DISTRIBUTION
-
The central limit theorem assures a normal (bell-shaped) distribution for a variable that is the summation of many independent, random inputs. This applies to single or multiple variables.
- COROLLA
-
Collectively, the petals of a flower.
- NECTAR GUIDES
-
Markings on the petals of flowers, often in contrasting colours or visible only in ultraviolet wavelengths, thought to act as directional beacons for pollinators, especially bees.
- ANTHER
-
The pollen-bearing part of the male floral structure (stamen).
- STIGMA
-
The region, usually the apex, of the gynoecium that receives pollen grains and on which the pollen germinates. The gynoecium is the seed-bearing organ of flowering plants, consisting of the stigma, style and ovary.
- STYLAR TUBE
-
In the genus Iris, the styles (tubular columns of tissue arising from the top of the ovary) look like flower petals and are tightly appressed to the top of the actual petals, forming this tube between the petal and the style.
- PHENOLOGY
-
The timing of periodic biological phenomena that are usually correlated with climactic conditions.
- HYBRID ZONE
-
A region of reproduction between individuals of different species, usually occurring where the ranges of the species come together.
- LIKELIHOOD-RATIO TEST STATISTIC
-
A maximum-likelihood method of hypothesis testing. The likelihood-ratio test statistic is twice the natural logarithm of the ratio of the maximum likelihood that the data fit the alternative hypothesis to the maximum likelihood that the data fit the null hypothesis.
- VARIANCE
-
A statistic that quantifies the dispersion of data about the mean. In quantitative genetics, the phenotypic variance (Vp) is the observed variation of a trait in a population. Vp can be partitioned into components, owing to genetic variance (Vg), environmental variance (Ve) and gene-by-environment correlations and interactions.
- SYMPATRIC
-
Occurring in the same area without loss of identity from interbreeding.
- MACROEVOLUTION
-
Evolution at or above the level of species.
- INTROGRESSIVE HYBRIDIZATION
-
Incorporation of genes from one species into the gene pool of another species.
- DEHISCENCE
-
The splitting open of a fruit.
- LOD SCORE
-
(Base 10 'logarithm of the odds', or 'log-odds'.) A method of hypothesis testing. The logarithm of the ratio between likelihoods under the null and alternative hypotheses.
- MONTE CARLO SIMULATION
-
The use of randomly generated or sampled data and computer simulations to obtain approximate solutions to complex mathematical and statistical problems.
- PERMUTATION TEST
-
A method of hypothesis testing. In these tests, an empirical distribution of a test statistic is obtained by permuting the original sample many times. Each permuted sample is considered to be a sample of the population under the null hypothesis.
- BAYESIAN APPROACH
-
An alternative statistical method that allows the use of prior information to evaluate the posterior probabilities of different hypotheses.
- SIMPLEX SEGREGATION
-
Segregation in polyploids. Segregation with no crossovers of the simplex genotype Aaaa would result in a gametic ratio of 1/2 Aa and 1/2 aa.
Rights and permissions
About this article
Cite this article
Mauricio, R. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat Rev Genet 2, 370–381 (2001). https://doi.org/10.1038/35072085
Issue Date:
DOI: https://doi.org/10.1038/35072085