Abstract
Living organisms adapt to light–dark rhythmicity using a complex programme based on internal clocks. These circadian clocks, which are regulated by the environment, direct various physiological functions. As the molecular mechanisms that govern clock function are unravelled, we are starting to appreciate simple patterns as well as exquisite layers of regulation.
Key Points
-
Living organisms adapt to light–dark rhythmicity using a complex programme based on internal clocks.
-
In mammals, the central clock structure is located within the suprachiasmatic nucleus (SCN) of the anterior hypothalamus.
-
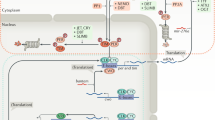
The clock can be considered as consisting of three overlapping components: the input pathways, the oscillator or pacemaker (which generates rhythmicity autonomously) and the output pathways.
-
Input-gene products are thought to sense external stimuli and relay the message to the oscillator to reset or entrain it.
-
Many pacemaker genes encode transcription factors or proteins that act on gene regulation, emphasizing the idea that the generation and modulation of rhythms relies mainly on transcriptional feedback loops and on activation and repression of gene expression (perhaps through chromatin remodelling).
-
The circadian clock probably uses a number of inter- or intragenic loops organized in networks.
-
Degradation of clock messenger RNAs and proteins is crucial to the control of oscillator periodicity. Entry of clock proteins into the nucleus is another checkpoint for circadian feedback loops.
-
Peripheral clocks may be controlled or synchronized by the SCN through blood-borne signalling factors oscillating in a circadian fashion.
-
Output gene-products, which include peptides and transcription factors, convey rhythmic information to downstream physiological systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Menaker, M., Moreira, L. F. & Tosini, G. Evolution of circadian organization in vertebrates. Braz. J. Med. Biol. Res. 30, 305–313 (1997).
Pittendrigh, C. S. Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54 (1993).An autobiographical essay by one of the fathers of modern chronobiology, covering the discovery of the basic principles of the discipline over fifty years.
Hastings, M. H. Central clocking. Trends Neurosci. 20, 459 –464 (1997).
Moore, R. Y. & Silver, R. Suprachiasmatic nucleus organization . Chronobiol. Int. 15, 475– 487 (1998).
Dunlap, J. C. Molecular bases for circadian clocks. Cell 96, 271–290 (1999).A comprehensive review on the genetics of the circadian clocks in cyanobacteria, fungi, Drosophila , mammals and plants.
Konopka, R. J. & Benzer, S. Clock mutants of Drosophila melanogaster . Proc. Natl Acad. Sci. USA 68, 2112 –2116 (1971).
King, D. P. et al. Positional cloning of the mouse circadian Clock gene . Cell 89, 641–653 (1997).The conclusion of an impressive study that led to the identification of the first mammalian clock gene, named Clock.
Lowrey, P. L. et al. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492 (2000).
Hogenesch, J. B., Gu, Y. Z., Jain, S. & Bradfield, C. A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl Acad. Sci. USA 95, 5474 –5479 (1998).
Gekakis, N. et al. Role of the CLOCK protein in the mammalian circadian mechanism . Science 280, 1564–1569 (1998).A key paper about a basic device, the feedback loop: the positive elements of the loop were finally uncovered.
Emery, P., So, W. V., Kaneko, M., Hall, J. C. & Rosbash, M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity . Cell 95, 669–679 (1998).Circadian photoreception in Drosophila is the duty of cryptochrome, a homologue of light-receiving proteins in plants.
Stanewsky, R. et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681 –692 (1998).
Foster, R. G. & Lucas, R. J. Clocks, criteria and critical genes . Nature Genet. 22, 217– 219 (1999).
Darlington, T. K. et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603 (1998).
Kume, K. et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 (1999).
Lee, C., Bae, K. & Edery, I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19, 5316 –5325 (1999).
Jin, X. et al. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57– 68 (1999).This was the first paper establishing a molecular link between the central clockworks and the expression of an output gene.
Ceriani, M. F. et al. Light-dependent sequestration of TIMELESS by CRYPTOCHROME . Science 285, 553–556 (1999).
Rothenfluh, A., Young, M. W. & Saez, L. A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 26, 505–514 (2000).
Griffin, E. A. Jr, Staknis, D. & Weitz, C. J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 (1999).
Shearman, L. P. et al. Interacting molecular loops in the mammalian circadian clock . Science 288, 1013–1019 (2000).
Yagita, K. et al. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 14, 1353– 1363 (2000).
van der Horst, G. T. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627– 630 (1999).
Cermakian, N., Whitmore, D., Foulkes, N. S. & Sassone-Corsi, P. Asynchronous oscillations of two zebrafish CLOCK partners reveal differential clock control and function. Proc. Natl Acad. Sci. USA 97, 4339–4344 (2000).
Hogenesch, J. B. et al. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J. Neurosci. 20, 1–5 (2000 ).
Gotter, A. L. et al. A time-less function for mouse timeless. Nature Neurosci. 3, 755–756 ( 2000).
Glossop, N. R., Lyons, L. C. & Hardin, P. E. Interlocked feedback loops within the Drosophila circadian oscillator. Science 286, 766–768 (1999).
Bae, K., Lee, C., Sidote, D., Chuang, K. Y. & Edery, I. Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol. 18, 6142–6151 (1998).
Oishi, K., Sakamoto, K., Okada, T., Nagase, T. & Ishida, N. Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem. Biophys. Res. Commun. 253 , 199–203 (1998).
Bae, K., Lee, C., Hardin, P. E. & Edery, I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 20, 1746–1753 (2000).
Lee, K., Loros, J. J. & Dunlap, J. C. Interconnected feedback loops in the Neurospora circadian system. Science 289, 107– 110 (2000).
Lucas, R. J. & Foster, R. G. Circadian clocks: A cry in the dark? Curr. Biol. 9, 825– 828 (1999).
Park, J. H. et al. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl Acad. Sci. USA 97, 3608–3613 (2000).
Gotter, A. L., Levine, J. D. & Reppert, S. M. Sex-linked period genes in the silkmoth, Antheraea pernyi: implications for circadian clock regulation and the evolution of sex chromosomes. Neuron 24, 953– 965 (1999).
Ding, J. M. et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713–1717 (1994).
Kornhauser, J. M., Nelson, D. E., Mayo, K. E. & Takahashi, J. S. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron 5, 127– 134 (1990).
Morris, M. E., Viswanathan, N., Kuhlman, S., Davis, F. C. & Weitz, C. J. A screen for genes induced in the suprachiasmatic nucleus by light. Science 279, 1544–1547 (1998).
Albrecht, U., Sun, Z. S., Eichele, G. & Lee, C. C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055– 1064 (1997).
Shearman, L. P., Zylka, M. J., Weaver, D. R., Kolakowski, L. F. Jr & Reppert, S. M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261– 1269 (1997).
Akiyama, M. et al. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J. Neurosci. 19, 1115– 1121 (1999).
Wollnik, F. et al. Block of c-Fos and JunB expression by antisense oligonucleotides inhibits light-induced phase shifts of the mammalian circadian clock. Eur. J. Neurosci. 7, 388–393 (1995).
Ginty, D. D. et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260 , 238–241 (1993).
Obrietan, K., Impey, S., Smith, D., Athos, J. & Storm, D. R. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J. Biol. Chem. 274, 17748–17756 (1999).
Belvin, M. P., Zhou, H. & Yin, J. C. The Drosophila dCREB2 gene affects the circadian clock. Neuron 22, 777–787 (1999).
de Cesare, D., Fimia, G. M. & Sassone-Corsi, P. Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem. Sci. 24, 281 –285 (1999).
Obrietan, K., Impey, S. & Storm, D. R. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nature Neurosci. 1, 693–700 (1998).
Akashi, M. & Nishida, E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645–649 (2000).
Ding, J. M. et al. A neuronal ryanodine receptor mediates light-induced phase delays of the circadian clock. Nature 394, 381–384 (1998).
Hastings, M. H. et al. Non-photic signalling in the suprachiasmatic nucleus. Biol. Cell 89, 495–503 ( 1997).
Maywood, E. S., Mrosovsky, N., Field, M. D. & Hastings, M. H. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc. Natl Acad. Sci. USA 96 , 15211–15216 (1999).
Shinohara, K., Hiruma, H., Funabashi, T. & Kimura, F. GABAergic modulation of gap junction communication in slice cultures of the rat suprachiasmatic nucleus. Neuroscience 96, 591–596 (2000).
Welsh, D. K., Logothetis, D. E., Meister, M. & Reppert, S. M. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995).
Pickard, G. E. & Rea, M. A. Serotonergic innervation of the hypothalamic suprachiasmatic nucleus and photic regulation of circadian rhythms. Biol. Cell 89, 513– 523 (1997).
Liu, C. et al. Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron 19, 91–102 (1997).
Liu, C. & Reppert, S. M. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 25, 123–128 (2000).
Cheung, P., Allis, C. D. & Sassone–Corsi, P. Signaling to chromatin through histone modifications. Cell (in the press).
Workman, J. L. & Kingston, R. E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67, 545–579 (1998).
Ko, H. P., Okino, S. T., Ma, Q. & Whitlock, J. P. Jr Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol. Cell. Biol. 16, 430–436 (1996).
Kobayashi, A., Numayama-Tsuruta, K., Sogawa, K. & Fujii-Kuriyama, Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt). J. Biochem. 122, 703–710 (1997).
Yao, T. P., Ku, G., Zhou, N., Scully, R. & Livingston, D. M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl Acad. Sci. USA 93, 10626–10631 (1996).
de Cesare, D., Jacquot, S., Hanauer, A. & Sassone-Corsi, P. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc. Natl Acad. Sci. USA 95, 12202–12207 ( 1998).
Sassone-Corsi, P. et al. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285, 886–891 (1999).
Dembinska, M. E., Stanewsky, R., Hall, J. C. & Rosbash, M. Circadian cycling of a PERIOD-beta-galactosidase fusion protein in Drosophila : evidence for cyclical degradation. J. Biol. Rhythms 12, 157–172 (1997).
So, W. V. & Rosbash, M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16, 7146–7155 ( 1997).
Ruoff, P., Vinsjevik, M., Monnerjahn, C. & Rensing, L. The Goodwin oscillator: on the importance of degradation reactions in the circadian clock. J. Biol. Rhythms 14, 469 –479 (1999).
Price, J. L. et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94 , 83–95 (1998). This was the first paper to identify a regulatory protein involved in the modification of other clock proteins.
Liu, Y., Loros, J. & Dunlap, J. C. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl Acad. Sci. USA 97, 234–239 (2000).
Talora, C., Franchi, L., Linden, H., Ballario, P. & Macino, G. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 18, 4961– 4968 (1999).
Nelson, D. C., Lasswell, J., Rogg, L. E., Cohen, M. A. & Bartel, B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340 ( 2000).
Somers, D. E., Schultz, T. F., Milnamow, M. & Kay, S. A. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis . Cell 101, 319–329 (2000).
Vielhaber, E., Eide, E., Rivers, A., Gao, Z. H. & Virshup, D. M. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20, 4888–4899 ( 2000).
Okamura, H. et al. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286, 2531–2534 (1999).
Vitaterna, M. H. et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl Acad. Sci. USA 96, 12114–12119 (1999).
Foster, R. G. Shedding light on the biological clock. Neuron 20, 829–832 (1998).
Freedman, M. S. et al. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science 284, 502– 504 (1999).Along with reference 76, this represented a big leap forward in the search for the mammalian circadian photoreceptor: it is neither the cones nor the rods, and so lies elsewhere in the retina.
Lucas, R. J., Freedman, M. S., Munoz, M., Garcia-Fernandez, J. M. & Foster, R. G. Regulation of the mammalian pineal by non-rod, non-cone, ocular photoreceptors. Science 284, 505–507 ( 1999).
Provencio, I. & Foster, R. G. Circadian rhythms in mice can be regulated by photoreceptors with cone-like characteristics. Brain Res. 694, 183–190 (1995).
Takahashi, J. S., DeCoursey, P. J., Bauman, L. & Menaker, M. Spectral sensitivity of a novel photoreceptive system mediating entrainment of mammalian circadian rhythms. Nature 308, 186–188 (1984).
Kojima, D., Mano, H. & Fukada, Y. Vertebrate ancient-long opsin: a green-sensitive photoreceptive molecule present in zebrafish deep brain and retinal horizontal cells. J. Neurosci. 20, 2845–2851 (2000).
Soni, B. G., Philp, A. R., Foster, R. G. & Knox, B. E. Novel retinal photoreceptors. Nature 394, 27–28 (1998).
Provencio, I. et al. A novel human opsin in the inner retina. J. Neurosci. 20, 600–605 ( 2000).
David-Gray, Z. K., Janssen, J. W., DeGrip, W. J., Nevo, E. & Foster, R. G. Light detection in a ‘blind’ mammal. Nature Neurosci. 1, 655– 656 (1998).
Whitmore, D., Foulkes, N. S. & Sassone-Corsi, P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404, 87–91 (2000).The striking message of this paper is that the peripheral organs and cells of zebrafish not only behave as independent clocks, but they have all the machinery to receive light and interpret these signals.
Wade, P. D., Taylor, J. & Siekevitz, P. Mammalian cerebral cortical tissue responds to low-intensity visible light. Proc. Natl Acad. Sci. USA 85, 9322–9366 (1988).
LeSauter, J. & Silver, R. Output signals of the SCN. Chronobiol. Int. 15, 535–550 (1998).
Ueyama, T. et al. Suprachiasmatic nucleus: a central autonomic clock. Nature Neurosci. 2, 1051–1053 (1999).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929– 937 (1998).This paper reports the first case of circadian oscillations in non-neuronal mammalian cells in culture.
Blau, J. & Young, M. W. Cycling vrille expression is required for a functional Drosophila clock. Cell 99, 661–671 (1999).
Ripperger, J. A., Shearman, L. P., Reppert, S. M. & Schibler, U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 (2000).
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C. & Taghert, P. H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791– 802 (1999).
Sarov-Blat, L., So, W. V., Liu, L. & Rosbash, M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 101, 647– 656 (2000).
Mueller, C. R., Maire, P. & Schibler, U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally . Cell 61, 279–291 (1990).
Lopez-Molina, L., Conquet, F., Dubois-Dauphin, M. & Schibler, U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 16, 6762–6771 (1997).
Yamaguchi, S. et al. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 20, 4773–4781 (2000).
Foulkes, N. S., Borjigin, J., Snyder, S. H. & Sassone-Corsi, P. Rhythmic transcription: the molecular basis of circadian melatonin synthesis . Trends Neurosci. 20, 487– 492 (1997).
Andretic, R., Chaney, S. & Hirsh, J. Requirement of circadian genes for cocaine sensitization in Drosophila. Science 285, 1066– 1068 (1999).
Kuhlman, S. J., Quintero, J. E. & McMahon, D. G. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport 11, 1479–1482 (2000).
Liu, C., Weaver, D. R., Strogatz, S. H. & Reppert, S. M. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell 91, 855–860 (1997).
Tosini, G. & Menaker, M. Circadian rhythms in cultured mammalian retina. Science 272, 419– 421 (1996).
Plautz, J. D., Kaneko, M., Hall, J. C. & Kay, S. A. Independent photoreceptive circadian clocks throughout Drosophila. Science 278, 1632–1635 (1997). This paper presents the first description of independent non-neural peripheral clocks in animals.
Giebultowicz, J. M., Stanewsky, R., Hall, J. C. & Hege, D. M. Transplanted Drosophila excretory tubules maintain circadian clock cycling out of phase with the host. Curr. Biol. 10, 107–110 (2000).
Whitmore, D., Foulkes, N. S., Strahle, U. & Sassone-Corsi, P. Zebrafish Clock rhythmic expression reveals independent peripheral circadian oscillators. Nature Neurosci. 1, 701–707 (1998).
Yamazaki, S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682– 685 (2000).
Krishnan, B., Dryer, S. E. & Hardin, P. E. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature 400, 375– 378 (1999).
Sakamoto, K. et al. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J. Biol. Chem. 273, 27039–27042 (1998).
Acknowledgements
We wish to apologize to all colleagues whose work, because of lack of space, could not be cited. We thank all the members of the Sassone-Corsi laboratory for helpful discussions. N.C. was supported by a Human Frontier Science Program Organization long-term fellowship and a Canadian Institutes of Health Research postdoctoral fellowship. Work in our laboratory is supported by grants from CNRS, INSERM, CHUR, Human Frontier Science Program, Organon Akzo/Nobel, Fondation pour la Recherche Médicale and Association pour la Recherche sur le Cancer.
Author information
Authors and Affiliations
Corresponding author
Glossary
- E-BOX ELEMENT
-
Promoter element recognized by transcription factors of the basic helix–loop–helix class.
- PHASE SHIFT
-
A shift in the endogenous circadian rhythms that occurs when an organism is placed in different lighting regimes (or other external stimuli).
- SUBJECTIVE NIGHT AND DAY
-
Correspond to the endogenous night–day circadian cycle of an organism, independent of the astronomical day and night.
- HISTONE ACETYL-TRANSFERASES AND DEACETYLASES
-
Enzymes that modify histones by adding and removing acetyl groups, a chemical modification thought to remodel chromatin structure.
- UBIQUITIN LIGASE
-
An enzyme that couples the small protein ubiquitin to lysine residues on a target protein, marking that protein for destruction by the proteasome.
- PHOTOLYASE
-
A DNA repair enzyme that splits pyrimidine dimers (lesions caused by UV irradiation) into monomers.
- OPSINS
-
Hydrophobic glycoproteins found in the visual pigments of vertebrates.
Rights and permissions
About this article
Cite this article
Cermakian, N., Sassone-Corsi, P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol 1, 59–67 (2000). https://doi.org/10.1038/35036078
Issue Date:
DOI: https://doi.org/10.1038/35036078
This article is cited by
-
Circadian rhythm modulates endochondral bone formation via MTR1/AMPKβ1/BMAL1 signaling axis
Cell Death & Differentiation (2022)
-
Circadian control of the secretory pathway maintains collagen homeostasis
Nature Cell Biology (2020)
-
Hippocampal clock regulates memory retrieval via Dopamine and PKA-induced GluA1 phosphorylation
Nature Communications (2019)
-
NSP-C contributes to the upregulation of CLOCK/BMAL1-mediated transcription
Cytotechnology (2019)
-
Disrupted circadian clocks and altered tissue mechanics in primary human breast tumours
Breast Cancer Research (2018)