Abstract
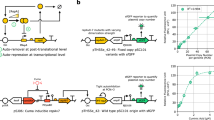
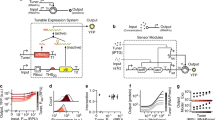
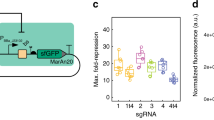
It has been proposed1 that gene-regulatory circuits with virtually any desired property can be constructed from networks of simple regulatory elements. These properties, which include multistability and oscillations, have been found in specialized gene circuits such as the bacteriophage λ switch2 and the Cyanobacteria circadian oscillator3. However, these behaviours have not been demonstrated in networks of non-specialized regulatory components. Here we present the construction of a genetic toggle switch—a synthetic, bistable gene-regulatory network—in Escherichia coli and provide a simple theory that predicts the conditions necessary for bistability. The toggle is constructed from any two repressible promoters arranged in a mutually inhibitory network. It is flipped between stable states using transient chemical or thermal induction and exhibits a nearly ideal switching threshold. As a practical device, the toggle switch forms a synthetic, addressable cellular memory unit and has implications for biotechnology, biocomputing and gene therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Modod, J. & Jacob, F. General conclusions: teleonomic mechanisms in cellular metabolism, growth and differentiation. Cold Spring Harb. Symp. Quant. Biol. 26, 389–401 (1961).
Ptashne, M. A Genetic Switch: Phage λ and Higher Organisms (Cell, Cambridge, Massachusetts, 1992).
Ishiura, M. et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 (1998).
Schellenberger, W., Eschrich, K. & Hofmann, E. Self-organization of a glycolytic reconstituted enzyme system: alternate stable stationary states, hysteretic transitions and stabilization of the energy charge. Adv. Enzyme Regul. 19, 257–284 (1980).
Glass, L. & Kauffman, S. A. The logical analysis of continuous, non-linear biochemical control networks. J. Theor. Biol. 39, 103–129 (1973).
Glass, L. Classification of biological networks by their qualitative dynamics. J. Theor. Biol. 54, 85–107 (1975).
Glass, L. Combinatorial and topological methods in nonlinear chemical kinetics. J. Chem. Phys. 63, 1325–1335 (1975).
Kauffman, S. The large scale structure and dynamics of gene control circuits: an ensemble approach. J. Theor. Biol. 44, 167– 190 (1974).
Thomas, R. Logical analysis of systems comprising feedback loops. J. Theor. Biol. 73, 631–656 ( 1978).
Thomas, R. Regulatory networks seen as asynchronous automata: a logical description. J. Theor. Biol. 153, 1– 23 (1991).
Tchuraev, R. N. A new method for the analysis of the dynamics of the molecular genetic control systems. I. Description of the method of generalized threshold models. J. Theor. Biol. 151, 71–87 (1991).
Arkin, A. & Ross, J. Computational functions in biochemical reaction networks. Biophys. J. 67, 560– 578 (1994).
Bhalla, U. S. & Iyengar, R. Emergent properties of networks of biological signaling pathways. Science 283, 381–387 (1999).
Yagilo, G. & Yagil, E. On the relation between effector concentration and the rate of induced enzyme synthesis. Biophys. J. 11, 11–27 (1971).
Shea, M. A. & Ackers, G. K. The OR control system of bacteriophage Lambda: a physical-chemical model for gene regulation. J. Mol. Biol. 181, 211–230 (1985).
Smith, T. F., Sadler, J. R. & Goad, W. Statistical–mechanical modeling of a regulatory protein: the Lactose repressor. Math. Biosci. 36, 61–86 (1977).
Arkin, A., Ross, J. & McAdams, H. H. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149, 1633–1648 (1998).
McAdams, H. H. & Arkin, A. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA 94, 814–819 (1997).
McAdams, H. H. & Arkin, A. Stimulation of prokaryotic genetic circuits. Annu. Rev. Biophys. Biomol. Struct. 27, 199–224 (1998).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Cormack, B. P., Valdivia, R. H. & Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996).
Ausubel, F. M. et al. Current Protocols in Molecular Biology (Wiley, New York, 1987).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Plainview, New York, 1989).
Edelstein-Keshet, L. Mathematical Models in Biology (McGraw-Hill, New York, 1988).
Kaplan, D. & Glass, L. Understanding Nonlinear Dynamics (Springer, New York, 1995).
Yagil, E. & Yagil, G. On the relation between effector concentration and the rate of induced enzyme synthesis. Biophys. J. 11, 11–27 (1971).
Rubinow, S. I. Introduction to Mathematical Biology (Wiley, New York, 1975).
Acknowledgements
We thank M. Bitensky and T. Yoshida for providing access to their flow cytometer; Y. Yu for his suggestions on plasmid construction; C. Sabanayagam for his technical advice; and C. Chow for his mathematical advice. This work was supported by the Office of Naval Research and the College of Engineering at Boston University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Center for Advanced Biotechnology, Boston University, 44 Cummington Street, Boston, Massachusetts 02215, USA
Supplementary information
Rights and permissions
About this article
Cite this article
Gardner, T., Cantor, C. & Collins, J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000). https://doi.org/10.1038/35002131
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35002131
This article is cited by
-
A computational model of stem cells’ internal mechanism to recapitulate spatial patterning and maintain the self-organized pattern in the homeostasis state
Scientific Reports (2024)
-
Engineered live bacteria as disease detection and diagnosis tools
Journal of Biological Engineering (2023)
-
Spatial biology of Ising-like synthetic genetic networks
BMC Biology (2023)
-
Multidimensional characterization of inducible promoters and a highly light-sensitive LOV-transcription factor
Nature Communications (2023)
-
Harnessing synthetic biology for advancing RNA therapeutics and vaccine design
npj Systems Biology and Applications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.