Abstract
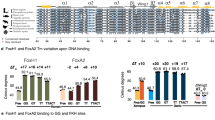
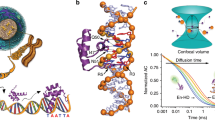
ZINC fingers1,2 constitute important eukaryotic DNA-binding domains, being present in many transcription factors3–5. The Cys2/His2zinc-finger class has conserved motifs of 28–30 amino acids which are usually present as tandem repeats1,2. The structure of a Cys2/His2zinc finger has been determined by nuclear magnetic resonance6, but details of its interaction with DNA were not established. Here we identify amino acids governing DNA-binding specificity using in vitro directed mutagenesis guided by similarities between the zinc fingers of transcription factors Spl (ref. 7) and Krox-20 (ref. 8). Krox-20 is a serum-inducible transcription activator8,9 which is possibly involved in the regulation of hindbrain development10; it contains three zinc fingers similar to those of Spl (refs 7, 8, 11) and binds to a 9-base-pair target sequence which is related to that of Spl (ref. 9). Our results show that each finger spans three nucleotides and indicate two positions in Krox-20 zinc fingers that are important for base-pair selectivity. Modelling with molecular graphics suggests that these residues could bind directly with the bases and that other amino acid–base contacts are also possible.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Miller, J., McLachlan, A. D. & Klug, A. EMBO J. 4, 1609–1614 (1985).
Brown, R. S., Sander, C. & Argos, P. FEBS Lett. 186, 271–274 (1985).
Hartshorne, T. A., Blumberg, H. & Young, E. T. Nature 320, 283–287 (1986).
Kadonaga, J. T., Courey, A. J., Ladika, J. & Tjian, R. Science 242, 1566–1570 (1988).
Vrana, K. E., Churchill, M. E. A., Tullius, T. D. & Brown, D. D. Molec. Cell. Biol. 8, 1684–1696 (1988).
Lee, M. S., Gippert, G. P., Soman, K. V., Case, D. A. & Wright, P. E. Science 245, 635–637 (1989).
Kadonaga, J., Carner, K. R., Maslarz, F. R. & Tjian, R. Cell 51, 1079–1090 (1987).
Chavrier, P. et al. EMBO J. 7, 29–35 (1988).
Chavrier, P. et al. EMBO J. 9, 1209–1218 (1990).
Wilkinson, D., Bhatt, S., Chavrier, P., Bravo, R. & Charnay, P. Nature 337, 461–464 (1989).
Chavrier, P. et al. Molec. Cell. Biol. 9, 787–797 (1989).
Gibson, T. J., Postma, J. P. M., Brown, R. S. & Argos, P. Protein Engng 2, 209–218 (1988).
Brown, R. S. & Argos, P. Nature 324, 215 (1986).
Berg, J. M. Proc. natn. Acad. Sci. U.S.A. 85, 99–102 (1988).
Fried, A. & Crothers, D. M. Nucleic Acids Res. 9, 6505–6525 (1981).
Strauss, F. & Varshavsky, A. Cell 37, 889–901 (1984).
Raymondjean, M., Cereghini, S. & Yaniv, M. Proc. natn. Acad. Sci. U.S.A. 85, 757–761.
Gessler, M. et al. Nature 343, 774–778 (1990).
Aggarwal, A. K., Rodgers, D. W., Drottar, M., Ptashne, M. & Harrison, S. C. Science 242, 899–907 (1988).
Jordan, S. R. & Pabo, C. O. Science 242, 893–899 (1988).
Seeman, N. C., Rosenberg, J. M. & Rich, A. Proc. natn. Acad. Sci. U.S.A. 73, 804–808 (1976).
Rhodes, D. & Klug, A. Cell 46, 123–132 (1986).
Berg, J. A. Rev. biophys. Chem. 19, 405–421 (1990).
Dayhoff, M. O. (ed.) Atlas of Protein Sequence and Structure (Natn. Biomed. Res. Fdn, Washington DC), 5, suppl. 3 (1978).
Saiki, R. K. et al. Science 239, 487–491 (1988).
Studier, F. W. & Moffatt, B. A. J. molec. Biol. 189, 113–130 (1986).
Rosenberg, A. H. et al. Gene 56, 125–135 (1987).
Towbin, H., Staehelin, T. & Gordon, J. Proc. natn. Acad. Sci. U.S.A. 76, 4350–4354 (1979).
Jones, T. A. J. appl. Crystallogr. 11, 268–272 (1978).
Dayring, H. E., Tramontano, A., Sprang, S. R. & Fletterick, R. J. J. Molec. Graph. 4, 82–87 (1986).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nardelli, J., Gibson, T., Vesque, C. et al. Base sequence discrimination by zinc-finger DNA-binding domains. Nature 349, 175–178 (1991). https://doi.org/10.1038/349175a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/349175a0
This article is cited by
-
Sequence variations of the EGR4 gene in Korean men with spermatogenesis impairment
BMC Medical Genetics (2017)
-
Dissection of a Krox20 positive feedback loop driving cell fate choices in hindbrain patterning
Molecular Systems Biology (2013)
-
Keep Your Fingers Off My DNA: Protein–Protein Interactions Mediated by C2H2 Zinc Finger Domains
Cell Biochemistry and Biophysics (2008)
-
The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment
Nature (2005)
-
EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK
Oncogene (2003)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.