Abstract
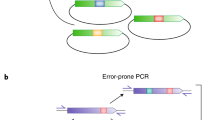
DNA shuffling is a powerful process for directed evolution, which generates diversity by recombination1,2, combining useful mutations from individual genes. Libraries of chimaeric genes can be generated by random fragmentation of a pool of related genes, followed by reassembly of the fragments in a self-priming polymerase reaction. Template switching causes crossovers in areas of sequence homology. Our previous studies used single genes and random point mutations as the source of diversity3,4,5,6. An alternative source of diversity is naturally occurring homologous genes, which provide ‘functional diversity’. To evaluate whether natural diversity could accelerate the evolution process, we compared the efficiency of obtaining moxalactamase activity from four cephalosporinase genes evolved separately with that from a mixed pool of the four genes. A single cycle of shuffling yielded eightfold improvements from the four separately evolved genes, versus a 270- to 540-fold improvement from the four genes shuffled together, a 50-fold increase per cycle of shuffling. The best clone contained eight segments from three of the four genes as well as 33 amino-acid point mutations. Molecular breeding by shuffling can efficiently mix sequences from different species, unlike traditional breeding techniques. The power of family shuffling may arise from sparse sampling of a larger portion of sequence space.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Stemmer, W. P. C. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA 91, 10747–10751 (1994).
Stemmer, W. P. C. Searching sequence space. Bio/Technology 13, 549–553 (1995).
Stemmer, W. P. C. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 (1994).
Zhang, J., Dawes, G. & Stemmer, W. P. C. Evolution of a fucosidase from a galactosidase by DNA shuffling and screening. Proc. Natl Acac. Sci. USA 94, 4504–4509 (1997).
Crameri, A., Whitehorn, E., Tate, E. & Stemmer, W. P. C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nature Biotech. 14, 315–319 (1996).
Crameri, A., Dawes, G., Rodriguez, E., Silver, S. & Stemmer, W. P. C. Molecular Evolution of an arsenate detoxification pathway by DNA shuffling. Nature Biotech. 15, 436–438 (1997).
Moore, J. C. & Arnold, F. H. Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nature Biotech. 14, 458–467 (1996).
Zhao, H. & Arnold, F. H. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 25, 1307–1308 (1997).
Lindberg, F. & Normark, S. Sequence of the Citrobacter freundii OS60 chromosomal ampC β-lactamase gene. Eur. J. Biochem. 156, 441–445 (1986).
Galleni, M. et al. Sequence and comparative analysis of three Enterobacter cloacae ampC β-lactamase genes and their products. Biochem. J. 250, 753–760 (1988).
Leiza, M. G. et al. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob. Agents Chemother. 38, 2150–2157 (1994).
Seoane, A., Francia, M. V. & Garcia Lobo, J. M. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 36, 1049–1052 (1992).
Stemmer, W. P. C., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. Single-step PCR assembly of a gene and a whole plasmid from large numbers of oligonucleotides. Gene 164, 49–53 (1995).
Lobkovsky, E. et al. Evolution of enzyme activity: crystallographic structure at 2 Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl Acad. Sci. USA 90, 11257–11261 (1993).
Kauffman, S. The Origins of Order (Oxford University Press, Oxford, (1993)).
Eigen, M. Steps Towards Life: a Perspective on Evolution (Oxford University Press, Oxford, (1992)).
Acknowledgements
We thank G. Dawes, J. Kieft, S. DelCardayre and M. Tobin and R. Howard, C.Yanofsky, P. Schultz, F. Arnold and A. Kornberg for useful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Crameri, A., Raillard, SA., Bermudez, E. et al. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 (1998). https://doi.org/10.1038/34663
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/34663
This article is cited by
-
Design of an artificial phage-display library based on a new scaffold improved for average stability of the randomized proteins
Scientific Reports (2023)
-
GH2 family β-galactosidases evolution using degenerate oligonucleotide gene shuffling
Biotechnology Letters (2023)
-
Systematic molecular evolution enables robust biomolecule discovery
Nature Methods (2022)
-
A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules
Nature Chemistry (2022)
-
A growth selection system for the directed evolution of amine-forming or converting enzymes
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.