Abstract
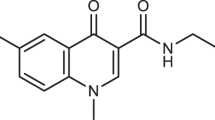
Ribonucleotide reductase, an essential enzyme for the synthesis of deoxyribonucleotides, is formed by the association of two non-identical subunits in almost all prokaryotic and eukaryotic cells1. The same model probably holds for the herpes simplex virus (HSV)-encoded ribonucleotide reductase2–6; two polypeptides of relative molecular mass 136,000 (136K; H1) and 40K (H2) (referred to elsewhere as RR1 and RR2; see for example, Dutia et al.26) have been associated with the viral enzyme by both genetic7,8 and immunological9–11 studies. Furthermore, DNA sequence analyses have shown significant stretches of amino-acid homology between these viral polypeptides and those of, respectively, subunit 1 (ref. 12) and subunit 2 (ref. 13) of the Escherichia coli and mammalian enzymes. To assess the involvement of the 40K polypeptide in reductase activity, we synthesized a nonapeptide corresponding to the sequence of its carboxy terminus with the intention of raising neutralizing antibodies specific for the viral activity (E.A.C. et al., in preparation). We report here the unexpected finding that the nonapeptide itself specifically inhibits the HSV ribonucleotide reductase activity in a reversible, non-competitive manner, and we suggest that it does this by impairment of the correct association of the two subunits. This phenomenon emphasizes the potential usefulness of synthetic peptides in probing critical sites involved in macromolecular interactions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Thelander, L. & Reichard, P. A. Rev. Biochem. 48, 133–158 (1979).
Ponce de Leon, M., Eisenberg, R. J. & Cohen, G. H. J. gen. Virol. 36, 163–173 (1977).
Huszar, D. & Bacchetti, S. J. Virol. 37, 580–588 (1981).
Langelier, Y. & Buttin, G. J. gen. Virol. 57, 21–31 (1981).
Averett, D. R., Lubbers, C., Elion, G. B. & Spector, T. J. biol. Chem. 258, 9831–9838 (1983).
Cohen, E. A., Charron, J., Perret, J. & Langelier, Y. J. gen. Virol. 66, 733–745 (1985).
Dutia, B. M. J. gen. Virol. 64, 513–521 (1983).
Preston, V. G., Palfreyman, J. W. & Dutia, B. M. J. gen. Virol. 65, 1457–1466 (1984).
Huszar, D., Beharry, S. & Bacchetti, S. J. gen. Virol. 64, 1327–1335 (1983).
Bacchetti, S., Evelegh, M. S., Muirhead, B., Sartori, C. S. & Huszar, D. J. Virol. 49, 591–593 (1984).
Frame, M. C., Marsden, H. S. & Dutia, B. M. J. gen. Virol. 66, 1581–1587 (1985).
Caras, I. W., Levinson, B. B., Fabry, M., Williams, S. R. & Martin, D. W. Jr J. biol. Chem. 260, 7015–7022 (1985).
Sjöberg, B. M. et al. FEBS Lett. 183, 99–102 (1985).
Draper, K. G., Frink, R. J. & Wagner, E. K. J. Virol. 43, 1123–1128 (1982).
Galloway, D. A. & Swain, M. A. J. Virol. 49, 724–730 (1984).
Merrifield, B. J. Am. chem. Soc. 85, 2149–2154 (1963).
Lewis, W. H. & Srinivasan, P. R. Molec. cell. Biol. 3, 1053–1061 (1983).
Summers, W. C. & Summers, W. P. J. Virol. 24, 314–318 (1977).
Weissbach, A., Hong, S.-C. L., Aucker, J. & Muller, R. J. biol. Chem. 248, 6270–6277 (1973).
Francke, B. & Garrett, B. Virology 116, 116–127 (1982).
Akiyama, S. K. & Yamada, K. M. J. biol. Chem. 260, 10402–10405 (1985).
Gartner, K. T. & Bennet, J. S. J. biol. Chem. 260, 11891–11894 (1985).
Ferreto, P., Guidotti, A., Conti-Tronconi, B. & Costa, E. Neuropharmacology 23, 1359 (1984).
Gillespie, L. L. et al. J. biol. Chem. 260, 16045–16048 (1985).
Benoit, R. et al. Proc. natn. Acad. Sci. U.S.A. 79, 917–921 (1982).
Dutia, B. M., Frame, M. C., Subak-Sharpe, J. H., Clark, W. N. & Marsden, H. S. Nature 321, 439–441 (1986).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cohen, E., Gaudreau, P., Brazeau, P. et al. Specific inhibition of herpesvirus ribonucleotide reductase by a nonapeptide derived from the carboxy terminus of subunit 2. Nature 321, 441–443 (1986). https://doi.org/10.1038/321441a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/321441a0
This article is cited by
-
TAS1553, a small molecule subunit interaction inhibitor of ribonucleotide reductase, exhibits antitumor activity by causing DNA replication stress
Communications Biology (2022)
-
Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets
Nature Reviews Drug Discovery (2003)
-
A potent peptidomimetic inhibitor of HSV ribonucleotide reductase with antiviral activity in vivo
Nature (1994)
-
Structure of ribonucleotide reductase protein R1
Nature (1994)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.