Abstract
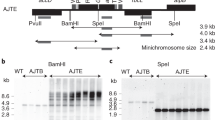
An almost universal feature of the circular chloroplast genome is a large inverted repeat sequence, some 10–25 kilobase pairs (kb) in size, which separates the remainder of the molecule into single copy regions of ∼80 kb and 20 kb1–3. A number of physical properties—formation of head-to-head dimers1, copy-correction between the inverted repeat segments1,3,4, resistance to intramolecular recombinational loss1–3, and maintenance of a highly stable chloroplast genome resistant to rearrangement2—have been attributed to the presence of this large inverted repeat. However, one property which an inverted repeat might be expected to confer—reversal of polarity of the single copy sequences located between the repeats1—has not yet been demonstrated for the chloroplast genome. I now show that chloroplast DNA prepared from a single plant of common bean (Phaseolus vulgaris) consists of two equimolar populations of molecules differing only in the relative orientation of their single copy sequences. A model is presented to explain these results, and comparisons are made to similar cases of inversion heterogeneity in 2-micrometre plasmid DNA from yeast5,6 and in herpes simplex virus DNA7,8.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Kolodner, R. & Tewari, K. K. Proc. natn. Acad. Sci. U.S.A. 76, 41–45 (1979).
Palmer, J. D. & Thompson, W. F. Cell 29, 537–550 (1982).
Gillham, N. W., Boynton, J. E. & Harris, E. H. in DNA Evolution: Natural Selection and Genome Size (ed. Cavalier-Smith) (Wiley, New York, in the press).
Gordon, K. H. J., Crouse, E. J., Bohnert, H. J. & Herrmann, R. G. Theor. appl. Genet. 61, 373–384 (1982).
Broach, J. R. Cell 28, 203–204 (1982).
Broach, J. R., Guarascio, V. R. & Jayaram, M. Cell 29, 227–234 (1982).
Roizman, B. Cell 16, 481–494 (1979).
Mocarski, E. S. & Roizman, B. Proc. natn. Acad. Sci. U.S.A. 78, 7047–7051 (1981).
Gordon, K. H. J., Crouse, E. J., Bohnert, H. J. & Herrmann, R. G. Theor. appl. Genet. 59, 281–296 (1981).
Palmer, J. D., Singh, G. P. & Pillay, D. T. N. Molec. gen. Genet. (in the press).
Mubumbila, M., Gordon, K. H. J., Crouse, E. J., Burkard, G. & Weil, J. H. Gene (submitted).
Kolodner, R. & Tewari, K. K. J. biol. Chem. 250, 8840–8847 (1975).
Koller, B. & Delius, H. Molec. gen. Genet. 178, 261–269 (1980).
Van Winkle-Swift, K. P. Curr. Genet. 1, 113–125 (1980).
Scowcroft, W. R. & Larkin, P. J. Theor. appl. Genet. 60, 179–184 (1981).
Palmer, J. D. & Stein, D. B. Curr. Genet. 5, 165–170 (1982).
Weststrate, M. W., Geelen, J.L.M.C. & Van der Noordaa, J. J. gen. Virol. 49, 1–21 (1980).
Chu, N. M., Oishi, K. K. & Tewari, K. K. Plasmid 6, 279–292 (1981).
Palmer, J. D. & Thompson, W. F. Proc. natn. Acad. Sci. U.S.A. 78, 5533–5537 (1981).
Zeig, J. & Simon, M. I. Proc. natn. Acad. Sci. U.S.A. 77, 4196–4200 (1980).
Bukhari, A. & Ambrosio, L. Nature 271, 575–577 (1978).
Palmer, J. D. thesis, Stanford Univ. (1981).
Palmer, J. D. Nucleic Acids Res. 10, 1593–1605 (1982).
Smith, G. E. & Summers, M. D. Analyt. Biochem. 109, 123–129 (1980).
Maniatis, T., Jeffrey, A. & Kleid, D. G. Proc. natn. Acad. Sci. U.S.A. 72, 1184–1188 (1975).
Palmer, J. D. & Thompson, W. F. Gene 15, 21–26 (1981).
Pogue-Geile, K. L., Dassarma, S., King, S. R. & Jaskunas, S. R. J. Bact. 142, 992–1003 (1980).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Palmer, J. Chloroplast DNA exists in two orientations. Nature 301, 92–93 (1983). https://doi.org/10.1038/301092a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/301092a0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.