Abstract
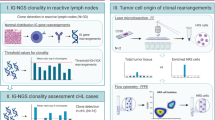
Lymphoproliferations are generally diagnosed via histomorphology and immunohistochemistry. Although mostly conclusive, occasionally the differential diagnosis between reactive lesions and malignant lymphomas is difficult. In such cases molecular clonality studies of immunoglobulin (Ig)/T-cell receptor (TCR) rearrangements can be useful. Here we address the issue of clonality assessment in 106 histologically defined reactive lesions, using the standardized BIOMED-2 Ig/TCR multiplex polymerase chain reaction (PCR) heteroduplex and GeneScan assays. Samples were reviewed nationally, except 10% random cases and cases with clonal results selected for additional international panel review. In total 75% (79/106) only showed polyclonal Ig/TCR targets (type I), whereas another 15% (16/106) represent probably polyclonal cases, with weak Ig/TCR (oligo)clonality in an otherwise polyclonal background (type II). Interestingly, in 10% (11/106) clear monoclonal Ig/TCR products were observed (types III/IV), which prompted further pathological review. Clonal cases included two missed lymphomas in national review and nine cases that could be explained as diagnostically difficult cases or probable lymphomas upon additional review. Our data show that the BIOMED-2 Ig/TCR multiplex PCR assays are very helpful in confirming the polyclonal character in the vast majority of reactive lesions. However, clonality detection in a minority should lead to detailed pathological review, including close interaction between pathologist and molecular biologist.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Van Dongen JJM, Langerak AW, Bruggemann M, Evans PAS, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Sandberg Y, Heule F, Lam K, Lugtenburg PJ, Wolvers-Tettero IL, van Dongen JJ et al. Molecular immunoglobulin/T- cell receptor clonality analysis in cutaneous lymphoproliferations. Experience with the BIOMED-2 standardized polymerase chain reaction protocol. Haematologica 2003; 88: 659–670.
Evans PA, Pott C, Groenen PJ, Salles G, Davi F, Berger F et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 2007, (in press).
Bruggemann M, White HE, Gaulard P, Garcia Sanz R, Gameiro P, Oeschger S et al. Powerful strategy for PCR-based clonality assessment in T-cell malignancies. Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia 2007, (in press).
Magro C, Crowson AN, Kovatich A, Burns F . Pityriasis lichenoides: a clonal T-cell lymphoproliferative disorder. Hum Pathol 2002; 33: 788–795.
Poenitz N, Dippel E, Klemke CD, Qadoumi M, Goerdt S . Jessner's lymphocytic infiltration of the skin: a CD8+ polyclonal reactive skin condition. Dermatology 2003; 207: 276–284.
Vanni R, Fletcher CD, Sciot R, Dal Cin P, De Wever I, Mandahl N et al. Cytogenetic evidence of clonality in cutaneous benign fibrous histiocytomas: a report of the CHAMP study group. Histopathology 2000; 37: 212–217.
De Vita S, De Re V, Gasparotto D, Ballare M, Pivetta B, Ferraccioli G et al. Oligoclonal non-neoplastic B cell expansion is the key feature of type II mixed cryoglobulinemia: clinical and molecular findings do not support a bone marrow pathologic diagnosis of indolent B cell lymphoma. Arthritis Rheum 2000; 43: 94–102.
Hanto DW, Birkenbach M, Frizzera G, Gajl-Peczalska KJ, Simmons RL, Schubach WH . Confirmation of the heterogeneity of posttransplant Epstein–Barr virus-associated B cell proliferations by immunoglobulin gene rearrangement analyses. Transplantation 1989; 47: 458–464.
Langerak AW, van Krieken JH, Wolvers-Tettero IL, Kerkhof E, Mulder AH, Vrints LW et al. The role of molecular analysis of immunoglobulin and T cell receptor gene rearrangements in the diagnosis of lymphoproliferative disorders. J Clin Pathol 2001; 54: 565–567.
Langerak AW, Sandberg Y, van Dongen JJ . Spectrum of T-large granular lymphocyte lymphoproliferations: ranging from expanded activated effector T cells to T-cell leukaemia. Br J Haematol 2003; 123: 561–562.
Sinclair E, Black D, Epling CL, Carvidi A, Josefowicz SZ, Bredt BM et al. CMV antigen-specific CD4+ and CD8+ T cell IFNgamma expression and proliferation responses in healthy CMV-seropositive individuals. Viral Immunol 2004; 17: 445–454.
Doisne JM, Urrutia A, Lacabaratz-Porret C, Goujard C, Meyer L, Chaix ML et al. CD8+ T cells specific for EBV, cytomegalovirus, and influenza virus are activated during primary HIV infection. J Immunol 2004; 173: 2410–2418.
van Leeuwen EM, Remmerswaal EB, Vossen MT, Rowshani AT, Wertheim-van Dillen PM, van Lier RA et al. Emergence of a CD4+CD28- granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J Immunol 2004; 173: 1834–1841.
Gamadia LE, van Leeuwen EM, Remmerswaal EB, Yong SL, Surachno S, Wertheim-van Dillen PM et al. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J Immunol 2004; 172: 6107–6114.
Katchar K, Wahlstrom J, Eklund A, Grunewald J . Highly activated T-cell receptor AV2S3(+) CD4(+) lung T-cell expansions in pulmonary sarcoidosis. Am J Respir Crit Care Med 2001; 163: 1540–1545.
Farzati B, Mazziotti G, Cuomo G, Ressa M, Sorvillo F, Amato G et al. Hashimoto's thyroiditis is associated with peripheral lymphocyte activation in patients with systemic sclerosis. Clin Exp Rheumatol 2005; 23: 43–49.
Gerber MA, Rapacz P, Kalter SS, Ballow M . Cell-mediated immunity in cat-scratch disease. J Allergy Clin Immunol 1986; 78 (5 Part 1): 887–890.
Chen W, Thoburn C, Hess AD . Characterization of the pathogenic autoreactive T cells in cyclosporine-induced syngeneic graft-versus-host disease. J Immunol 1998; 161: 7040–7046.
Chetty R, Vaithilingum M, Thejpal R . Epstein–Barr virus status and the histopathological changes of parotid gland lymphoid infiltrates in HIV-positive children. Pathology 1999; 31: 413–417.
Langerak AW, Szczepanski T, Van der Burg M, Wolvers-Tettero ILM, Van Dongen JJM . Heteroduplex PCR analysis of rearranged Tcell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997; 11: 2192–2199.
Kneba M, Bolz I, Linke B, Hiddemann W . Analysis of rearranged T-cell receptor beta-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood 1995; 86: 3930–3937.
Iijima T, Inadome Y, Noguchi M . Clonal proliferation of B lymphocytes in the germinal centers of human reactive lymph nodes: possibility of overdiagnosis of B cell clonal proliferation. Diagn Mol Pathol 2000; 9: 132–136.
Brauninger A, Yang W, Wacker HH, Rajewsky K, Kuppers R, Hansmann ML . B-cell development in progressively transformed germinal centers: similarities and differences compared with classical germinal centers and lymphocyte-predominant Hodgkin disease. Blood 2001; 97: 714–719.
Droese J, Langerak AW, Groenen PJ, Bruggemann M, Neumann P, Wolvers-Tettero IL et al. Validation of BIOMED-2 multiplex PCR tubes for detection of TCRB gene rearrangements in T-cell malignancies. Leukemia 2004; 18: 1531–1538.
Lee MW, Lee DP, Choi JH, Moon KC, Koh JK . Failure to detect clonality in eosinophilic pustular folliculitis with follicular mucinosis. Acta Derm Venereol 2004; 84: 305–307.
Acknowledgements
We thank Ingrid LM Wolvers-Tettero, Ellen J van Gastel-Mol, Monique ECM Oud, Brenda Verhaaf (Rotterdam), Francis Devez (Paris), Helen White (Southampton), Jonathan Langdown, Karen Brown (Cambridge), Patrícia Pontes (Porto), Kirsten Linsmeier, Nicola Passow, Cornelia Rütz, Carmen Scherer (Heidelberg), Klaas Kooistra and Jelle Conradie (Groningen) for technical assistance. The BIOMED-2 Pathology Review Panel consisted of JHJM van Krieken (Nijmegen, coordinator), F Berger (Lyon), B Jasani (Cardiff), D Canioni (Paris), PhM Kluin (Groningen), JM Cabeçadas (Lisboa), TJ Molina (Paris), G Delsol (Toulouse), A Orfao (Salamanca), JF Garcia (Madrid), M Ott (Würzburg), P Gaulard (Creteil), ST Pals (Amsterdam) and ML Hannsmann (Frankfurt). This study was supported by the BIOMED-2 Concerted Action grant BMH4-CT98-3936 of the European Commission. TJM was supported by a ‘Ligue contre le cancer, Comité de Paris’ equipment grant. FLL was supported by a grant from the Leukaemia Research Fund, UK.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Langerak, A., Molina, T., Lavender, F. et al. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls. A report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 21, 222–229 (2007). https://doi.org/10.1038/sj.leu.2404482
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2404482
Keywords
This article is cited by
-
Diagnostic and prognostic molecular pathology of lymphoid malignancies
Virchows Archiv (2023)
-
Evaluation of a worldwide EQA scheme for complex clonality analysis of clinical lymphoproliferative cases demonstrates a learning effect
Virchows Archiv (2021)
-
T-cell clones of uncertain significance are highly prevalent and show close resemblance to T-cell large granular lymphocytic leukemia. Implications for laboratory diagnostics
Modern Pathology (2020)
-
Comprehensive assessment of peripheral blood TCRβ repertoire in infectious mononucleosis and chronic active EBV infection patients
Annals of Hematology (2017)