Abstract
Clonality analysis in classic Hodgkin lymphoma (cHL) is of added value for correctly diagnosing patients with atypical presentation or histology reminiscent of T cell lymphoma, and for establishing the clonal relationship in patients with recurrent disease. However, such analysis has been hampered by the sparsity of malignant Hodgkin and Reed-Sternberg (HRS) cells in a background of reactive immune cells. Recently, the EuroClonality-NGS Working Group developed a novel next-generation sequencing (NGS)-based assay and bioinformatics platform (ARResT/Interrogate) to detect immunoglobulin (IG) gene rearrangements for clonality testing in B-cell lymphoproliferations. Here, we demonstrate the improved performance of IG-NGS compared to conventional BIOMED-2/EuroClonality analysis to detect clonal gene rearrangements in 16 well-characterized primary cHL cases within the IG heavy chain (IGH) and kappa light chain (IGK) loci. This was most obvious in formalin-fixed paraffin-embedded (FFPE) tissue specimens, where three times more clonal cases were detected with IG-NGS (9 cases) compared to BIOMED-2 (3 cases). In total, almost four times more clonal rearrangements were detected in FFPE with IG-NGS (N = 23) as compared to BIOMED-2/EuroClonality (N = 6) as judged on identical IGH and IGK targets. The same clonal rearrangements were also identified in paired fresh frozen cHL samples. To validate the neoplastic origin of the detected clonotypes, IG-NGS clonality analysis was performed on isolated HRS cells, demonstrating identical clonotypes as detected in cHL whole-tissue specimens. Interestingly, IG-NGS and HRS single-cell analysis after DEPArray™ digital sorting revealed rearrangement patterns and copy number variation profiles indicating clonal diversity and intratumoral heterogeneity in cHL. Our data demonstrate improved performance of NGS-based detection of IG gene rearrangements in cHL whole-tissue specimens, providing a sensitive molecular diagnostic assay for clonality assessment in Hodgkin lymphoma.
Similar content being viewed by others
Introduction
Classic Hodgkin lymphoma (cHL) is a B-cell neoplasm, characterized by the presence of a small number of malignant cells, called Hodgkin and Reed-Sternberg (HRS) cells, residing in a background of inflammatory cells, including reactive B cells and plasma cells1,2. In cHL, the HRS cells represent functionally impaired B lymphocytes that have lost the capacity to express immunoglobulins and many of the B-cell markers, but show CD15 and CD30 expression3. Evidence for their B-cell origin has been obtained by the detection of clonal rearrangements of the immunoglobulin genes, i.e., the immunoglobulin heavy (IGH) and light chain gene loci4,5,6,7. Furthermore, the presence of somatic hypermutation (SHM) within the rearranged variable (V) genes demonstrates that HRS cells originate from (post-)germinal center B lymphocytes8,9,10. Although most cHL cases are diagnosed based on clinical presentation and histopathology, the molecular detection of clonal immunoglobulin (IG) gene rearrangements is helpful for diagnosing cases with a more atypical presentation and a histology similar to T cell lymphoma, or for clonal comparison in patients with recurrent disease11,12.
Currently, the gold standard for clonality testing of lymphomas is the BIOMED-2/EuroClonality assay developed and validated by the EuroClonality Consortium13,14,15,16. For assessing B-cell clonality, multiplex PCR reactions are performed for the detection of IGH and kappa light chain (IGK) gene rearrangements, followed by fragment analysis using GeneScan or heteroduplex analysis. The applicability of the BIOMED-2/EuroClonality assay in cHL has been evaluated in different studies, demonstrating B-cell clonality in about 50% of formalin-fixed paraffin-embedded (FFPE) cHL tissue specimens7,17,18,19,20,21,22,23. Although the combination of both IGH and IGK rearrangement analysis, including incomplete rearrangements not undergoing SHM, has certainly enhanced the detection rate of clonal rearrangements in cHL, more sensitive techniques can improve clonality assessment in cHL.
Recently, a next-generation sequencing (NGS)-based clonality assay for detection of IG gene rearrangements (IG-NGS) has been developed by the EuroClonality-NGS Working Group24. In this assay, new primers have been designed for complementary IG gene targets that will result in shorter amplicon length, particularly for the incomplete IGH targets as well as IGK targets. This enables better clonality detection in genomic DNA from FFPE tissue specimens, especially samples with inferior DNA quality. Another advantage of IG-NGS is the immediate sequence availability of the identified clonotypes, both of the malignant clone as well as the non-neoplastic background B cells. This will provide more reliable information to detect minor clones in a background of polyclonal B cells and assessing the clonal relationship of multiple B-cell malignancies within the same patient.
In this study, we compared the performance of NGS-based clonality assessment in cHL to the conventional BIOMED-2/EuroClonality assay to identify clonal IGH and IGK gene rearrangements in whole-tissue specimens, including both FFPE and fresh frozen (FF) tissue. We demonstrate improved performance of the IG-NGS assay to detect B-cell clonality in both types of tissue specimens. Furthermore, our data reveal evidence for biclonality in some cHL tumor samples, indicating that co-existence of multiple malignant clones and clonal diversity can occur in this type of lymphoma.
Materials and methods
Selection of cHL and reactive lymphoid tissue samples
This study was performed with 16 lymph node biopsies of patients diagnosed with cHL according to the 2017 revised 4th edition of the World Health Organization classification25, for which both FFPE and FF tissue material was available18 (Table 1 and Supplementary Table S1). IG-NGS was analyzed on all 16 cases, both FFPE and FF, while BIOMED-2/EuroClonality analysis could only be performed on 15 FFPE cases, due to limited DNA yield for case 13, but included all 16 FF cases. For clone definition and identification, available IG-NGS clonality data of 30 reactive lymphoproliferative samples was analyzed (Supplementary Table S2)26. All cHL samples were retrieved from the local archive of the Department of Pathology at Radboud University Medical Center, and collected in accordance with the declaration of Helsinki. This study received approval of the institutional CMO review board (Approval No. 2020-6390).
Clonality assessment by next-generation sequencing
Detection of IGH and IGK gene rearrangements by next-generation sequencing (IG-NGS) was performed as recently described24. In short, three multiplex PCR reactions were performed with 40–50 ng input DNA for the detection of IG gene rearrangements, which involved variable (V), diversity (D), joining (J), kappa deleting element (KDE), and the recombination signal sequence (RSS) gene segments of the following five targets: IGHV-IGHD-IGHJ (FR3), IGHD-IGHJ, IGKV-IGKJ, IGKV-KDE, and Intron RSS-KDE. After bead purification, library preparation and amplification, the samples were pooled with equal DNA concentrations (corresponding to equimolar amounts) for sequencing on Ion Torrent PGM. The Ion 318™ Chip Kit v2 BC (Ion Torrent™, Thermo Fisher, Waltham, MA, USA) was loaded with 24 or 32 samples per chip. The sequencing results were analyzed and visualized using the bioinformatics pipeline ARResT/Interrogate (http://arrest.tools/interrogate)27. The output data defines a clonotype (e.g., V3-9 -1/20/-6 J3), notated as a combination of the 5′ gene (V3-9), nucleotides deleted from the 5′ gene (-1), the added nucleotides at the junction including the D-segment if present (20), nucleotides deleted from the 3′ gene (-6), and the 3′ gene (J3)28. Data were considered reliable with at least a minimum amount of 1000 reads for IGHV-IGHD-IGHJ (mean: 34,917 reads) and IGKV-IGKJ (mean: 12,032 reads), and 500 reads for IGHD-IGHJ (mean: 31,242 reads), IGKV-KDE (mean: 4201 reads), and Intron RSS-KDE (mean: 2869 reads). For the assessment of B-cell clonality, each DNA sample (extracted as independent isolates from FF and FFPE specimens of the same cHL tumor tissue) was analyzed in duplicate by multiplex PCR and NGS to rule out misinterpretation or artifacts, and validate less-abundant clonotypes. Only clonotypes that are detected with a dominant abundancy above threshold in duplicate are considered as clonal rearrangements.
Additional information regarding ‘Materials and Methods’, including the strategy for clone identification, enrichment of HRS cells and single-cell analysis are provided in the Supplementary information.
Results
Clonality detection of selected cHL specimens by conventional BIOMED-2/EuroClonality assay
In this study 16 classic Hodgkin lymphoma (cHL) cases were included, of which both archival FF and FFPE tissue specimens were available (Table 1 and Supplementary Table S1)18. Eleven cases were of the nodular sclerosing subtype (NSHL), of which four cases (36%) were Epstein-Barr virus (EBV)-positive, and five cases were diagnosed as mixed cellularity HL (MSHL), with two EBV-positive cases (40%). The average percentage of malignant HRS cells in total tissue as judged by CD30 immunohistochemical staining varied between 1% and 15%, and the fraction of HRS cells related to total B-cell counts ranged from 3% to 43% (Table 1 and Supplementary Fig. S3). First, conventional BIOMED-2/EuroClonality was performed on 15 FFPE cases, for which sufficient DNA was available, and 16 FF cases.. Clonal IG gene rearrangements for at least one of the IG targets were detected 10 out of 16 FF samples (63%) and 3 out of 15 FFPE samples (20%) (Table 2 and Supplementary Table S3). The remaining six FF samples showed a polyclonal pattern (34%), while this was observed in 10 out 15 FFPE samples (67%), of which seven displayed nonevaluable results on at least one of the IG targets. Two FFPE samples (case 3 and 10; 13%) yielded no interpretable results on all IG targets, due to the inferior DNA quality (Supplementary Table S3). Notably, in none of the 16 cases clonal IGKV-IGKJ gene rearrangements were detected with the BIOMED-2/EuroClonality assay, due to suboptimal detection of minor clonal rearrangements in the IGKV-IGKJ GeneScan pattern, which displayed polyclonal background over three narrow Gaussian curves.
Detection of clonal immunoglobulin gene rearrangements in cHL by IG-NGS
To define clonality in cHL by IG-NGS, we reasoned that it would be important to identify the clone frequencies of abundant clonotypes in non-malignant reactive lymphoproliferative samples, which would define thresholds above which potentially malignant clonotypes could be identified. Furthermore, it would be essential to demonstrate that identified clonotypes in cHL whole-tissue specimens corresponded to IG gene rearrangements of the corresponding HRS cells, and not reactive B-cell clones. To this end, we established a specific workflow procedure as summarized in Fig. 1.
For clone identification in classic Hodgkin lymphoma (cHL), first the relative frequencies of the most-abundant clonotypes in non-malignant reactive lymph nodes (N = 30) were determined by next-generation sequencing of immunoglobulin gene rearrangements (IG-NGS) (Supplementary Fig. S1). Here, clonality analysis was performed in duplicate, followed by calculation of the mean ratio of the top-4 overlapping clonotypes identified in duplicate in each sample, for each of the immunoglobulin heavy chain (IGH) and immunoglobulin kappa light chain (IGK) targets, including both complete and incomplete IG gene rearrangements. Based on the maximum ratio of the overlapping clonotypes in the reactive lymph nodes, a threshold value for clonality was defined for each of the targets. The established threshold values were used to define clonality in the 16 cHL cases. For clone validation, laser microdissection and flow cytometry were used to enrich the Hodgkin Reed-Sternberg (HRS) cells from the tumor-tissue specimen of two cHL cases, and these HRS cell fractions were subjected to IG-NGS clonality analysis to validate these as neoplastic clones.
First, the normal distribution of IG gene rearrangements for all five targets (framework-3 (FR3) IGHV-IGHD-IGHJ, IGHD-IGHJ, IGKV-IGKJ, IGKV-KDE, and Intron RSS-KDE) was established in a cohort of reactive lymphoid tissue specimens (N = 30; RLN1-RLN30) (Supplementary Table S2). Here, we focused on the detection of identical clonotypes in two independent IG-NGS assays and sequence runs derived from one reactive tissue sample. By calculating the mean ratio of the top 4 overlapping clonotypes compared to the background clonotypes, the relative abundances of the most dominant clonotypes were defined for each target (Supplementary Fig. S1). Secondly, threshold ratio values were determined for each target in the reactive lymphoproliferative samples by comparing frequencies of the abundant clonotypes relatively to the minor clonotypes (Supplementary Fig. S4). Threshold values for clonality were set as such that all reactive lymphoproliferative samples did not exceed this ratio, resulting in the values 2.5 for IGHV-IGHD-IGHJ (FR3), 4.5 for IGHD-IGHJ, 4 for IGKV-IGKJ, and 2.5 for IGKV-KDE clonotypes. Based on the limited diversity in Intron RSS-KDE rearrangements, the threshold was based on the relative mean abundance of all rearrangements involving KDE, and determined on 4.5% (no ratio calculation).
Next, NGS-based clonality assessment was performed on each of the 16 cHL cases in duplicate, for both FF and FFPE samples. Taking the established thresholds for clone definition, we could detect clonal rearrangements in at least one of the IG targets in 14 out of 16 FF cases (88%) and 9 out of 16 FFPE cases (56%) with IG-NGS (Table 2, Supplementary Tables S3 and S4, and Supplementary Fig. S5). FF yielded interpretable results for all samples, while FFPE remained problematic in two DNA samples (case 10 and 13), which yielded too low read counts for clone identification on multiple targets, due to inferior DNA quality (Supplementary Table S3). Identical clonal rearrangements were detected in the paired FFPE and FF tissues with clonal IG gene rearrangements, demonstrating a good correlation. Alignment of the obtained IGHV and IGKV nucleotide sequences of the identified cHL clonotypes to their germline sequences showed evidence for SHM within the malignant cHL clones (Supplementary Fig. S6).
To actually confirm that the identified clonotypes were of neoplastic cell origin, we enriched the HRS cells in FF and FFPE material of two cHL cases (case 8 and 16) using laser microdissection (LMD) of CD30+ HRS cells in FF tissue sections, and flow cytometric isolation of CD30+MUM1+ HRS cells after mild enzymatic digestion of the paired FFPE tissue specimen, as described by Juskevicius et al.29. Genomic DNA was isolated from the HRS-enriched cell fractions of both procedures, and subjected to IG-NGS analysis. In each of the two cHL cases, identical clonotypes were detected in higher abundance in the HRS-enriched cell fractions as compared to whole-tissue specimens, both for FF as well as FFPE tissues (Fig. 2 and Supplementary Table S5). These results confirmed that the clonal IG gene rearrangements and corresponding clonotypes as detected by IG-NGS in cHL are of neoplastic cell origin.
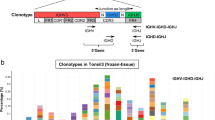
NGS-based clonality analysis was performed on DNA isolated from whole-tumor tissue and Hodgkin Reed-Sternberg (HRS)-enriched cell suspensions of a classic Hodgkin lymphoma (cHL) sample (case 16), archived as a fresh frozen (FF) and formalin-fixed paraffin-embedded (FFPE) tissue specimen. The isolation of HRS cells was performed with laser microdissection (LMD) of CD30+ cells in FF tissue, and flow cytometric sorting of CD30+/MUM1+ HRS cells after mild enzymatic digestion of FFPE tissue. Identified clonotypes for target IGHD-IGHJ of FF (A) and FFPE (B) specimen are shown, in the order whole tissue, followed by HRS-enriched cell fraction.
Improved performance of IG-NGS compared to BIOMED-2/EuroClonality assay for clonality assessment in cHL
To evaluate the actual performance of IG-NGS for clonality assessment in cHL, we compared the results of NGS-based clonality analysis with the BIOMED-2/EuroClonality assay (Fig. 3). Notably, with conventional BIOMED-2/EuroClonality analysis, detection of IGHV-IGHD-IGHJ gene rearrangements is based on primers covering FR1, FR2, and FR3 regions. Since in our current IG-NGS assay, no FR1 and FR2 primers were included for detecting IGHV-IGHD-IGHJ gene rearrangements, the comparison involved only the FR3 region. IG-NGS was able to detect clonal rearrangements in 14 cHL cases (88%) as compared to 10 cases (63%) with BIOMED-2/EuroClonality assay, when assessing FF DNA (Table 2). In 11 out of 14 clonal FF samples (79%), clonality was detected on multiple targets with IG-NGS, while this was observed in only 2 out of 10 clonal FF samples (20%) with the BIOMED-2/EuroClonality assay (Table 3 and Supplementary Table S3). In FF, IG-NGS detected clonal rearrangements in six additional samples for target IGHV-IGHD-IGHJ (FR3), two samples for IGHD-IGHJ, six samples for IGKV-IGKJ and three samples for IGKV/Intron RSS-KDE targets as compared to BIOMED-2/EuroClonality assay. Two samples (cases 9 and 13) showed also additional clonal IGKV/Intron RSS-KDE rearrangements with IG-NGS (Table 3), demonstrating the increased sensitivity and specificity of IG-NGS clonality testing in FF tissue. This is due to the improved detection of clonotypes within a polyclonal background, especially for IGKV-IGKJ and IGKV/intron RSS-KDE rearrangements30. However, IG-NGS failed to detect an IGHV-IGHD-IGHJ gene rearrangement for case 16, which was identified with conventional BIOMED-2/EuroClonality testing for FR3 using a different IGHV-FR3 forward primer (Table 3). Further evaluation of the FR1 BIOMED-2/GeneScan data of this case showed a shorter amplicon length than expected (173 bp vs ~230–295 bp) (Supplementary Fig. S7). Sanger sequencing of this amplicon revealed a 97 bp deletion within the IGHV3-74 gene, which harbored the annealing site of the IG-NGS FR3 primer, while the sequence of the BIOMED-2 FR3 primer was still present (Supplementary Fig. S7).
The results of clonality analysis for (A) fresh frozen (FF) and (B) formalin-fixed paraffin-embedded (FFPE) tissue of case 12 are shown for all IG targets (IGHV-IGHD-IGHJ, IGHD-IGHJ, IGKV-IGKJ, and IGKV/Intron RSS-KDE) based on next-generation sequencing of immunoglobulin gene rearrangements (IG-NGS) (left panels) and BIOMED-2/GeneScan assay (right panels). IG-NGS clonality results: junction length in amino acids (junction aa length) is shown on the x-axis, the abundancy of clonotypes is shown in percentages on the y-axis. Each bar represents a total of all detected clonotypes with the same junction length, each color represents a unique clonotype based on their nucleotide sequence. The 50 most-abundant clonotypes are shown in colors, all other clonotypes are depicted together in gray. Clonotypes are described by the 5′ and 3′ gene annotation and the junctional nucleotide sequence of the rearrangement. BIOMED-2/GeneScan clonality results: clonal product is marked with black arrow for IGKV/Intron RSS-KDE in FFPE sample; *non-specific IGHD-IGHJ non-specific germline product; #IGKV-IGKJ polyclonal signal, the narrow Gaussian curve in the IGKV 1f/6/7-IGKJ size window is due the high homology of the IGKV genes and explains why minor clonal rearrangements can be easily missed.
In FFPE tissue specimens, IG-NGS detected an even higher number of additional clonal IG gene rearrangements with the BIOMED-2/EuroClonality assay. In total, 9 cases (56%) showed clonal IG gene rearrangements with IG-NGS, while only 3 clonal cases (20%) were detected with BIOMED-2/EuroClonality, and clonality assessment with IG-NGS was often supported by multiple targets (Table 3 and Supplementary Table S3). Similar to the FF sample, one clonal IGHV-IGHD-IGHJ rearrangement could not be detected with IG-NGS, due to a 97 bp deletion (case 16). There was one cHL tumor (case 6) with a clonal IGHV-IGHD-IGHJ gene rearrangement detected in FR2 by the BIOMED-2/EuroClonality assay, which was not identified by the IG-NGS approach since FR2 was not included in the current assay (Table 3). Overall, only one (case 15) out of 16 cHL samples (6%) showed no detectable IG gene rearrangements with both techniques.
Detection of clonal diversity and intratumoral heterogeneity in cHL by single-cell analysis
Since NGS-based clonality assessment revealed more detailed information regarding the clonal composition of lymphomas, two out of 14 cases (cases 8 and 9; 14%) showed multiple clonal IG gene rearrangements exceeding the allelic capacity of a single B-cell clone. Pathology review provided no indications for a composite lymphoma in both cHL FFPE tissue specimens. Case 8 displayed two unproductive IGHD-IGHJ gene rearrangements (IGHD3-9/IGHJ6 and IGHD1-26/IGHJ5) combined with one productive IGHV-IGHD-IGHJ gene rearrangement (IGHV3-9/IGHD6-13/IGHJ3), implying the presence of two B-cell clones (Supplementary Fig. S8). All three IGH gene rearrangements were identified in both whole-tissue and bulk HRS-isolated cells (Supplementary Table S5). Case 9 showed an even more complex pattern of unrelated IG gene rearrangements. Multiple clonal rearrangements were detected on both IGH (two in-frame IGHV-IGHD-IGHJ rearrangements) and IGK targets (three unrelated IGKV-IGKJ and four IGKV/Intron RSS-KDE rearrangements), suggesting also biclonality (Table 3, Supplementary Table S4, and Fig. S8). These data demonstrate that clonal diversity in cHL can be determined by deep-sequencing of IG gene rearrangements.
To confirm in an independent manner the clonal composition of these two cHL cases, we aimed to perform copy number variation (CNV) profiling on CD30+PD-L1+ HRS single cells isolated from FFPE specimens by DEPArray™ sorting technology31,32. This procedure involves whole-genome amplification (WGA) of single cells followed by shallow whole-genome sequencing (sWGS), and subsequent CNV analysis of the individual HRS cells. DNA integrity analysis with quantitative PCR on DNA isolated from the FFPE tissue specimens indicated that only case 9 (QC score: 0.35) fulfilled the quality criteria for the subsequent sWGS approach. Unsupervised hierarchical cluster analysis revealed two separate clusters with distinct CNV profiles of the individual HRS cells (Fig. 4 and Supplementary Figs. S9 and S10). Each cluster showed defined regions with copy number gains and losses, and both populations showed evidence of intraclonal heterogeneity, reminiscent of the genomic instability described for cHL33,34. Overall, NGS-based clonality assessment showed improved performance in cHL, including increased sensitivity and specificity of detecting clonal IG gene rearrangements, and providing accurate information on the clonal composition of the lymphoid malignancy.
Single HRS cells, which were obtained after mild enzymatic digestion of formalin-fixed paraffin-embedded tissue specimen (case 9), were isolated with DEPArray digital sorting technology based on CD30 and PD-L1 expression. Subsequently, genomic DNA was isolated and subjected to whole-genome amplification, shallow whole-genome sequencing and copy number analysis. HRS cells are defined as hyperdiploid, CD30+ PD-L1+ enlarged cells/nuclei. A Microscopy images and corresponding copy number variation (CNV) profiles of two representative single HRS cells (one from each cluster) and control leukocyte of the same tissue. The CNV profiles show the genome location according to chromosome position on the x-axis and the log-scaled copy number values on the y-axis. B Unsupervised hierarchical clustering of CNV profiles of 30 HRS cells (21 single HRS cells (yellow) and 9 rosetted HRS cells (blue)) reveals two main clusters (designated with vertical lines), indicating biclonality of the cHL-tumor tissue. The x-axis shows the chromosome position and the y-axis the CNV profiles of the individual HRS cells.
Discussion
Detection of IG gene rearrangements for clonality analysis in cHL has remained challenging due to the low frequency of malignant HRS cells in a background of reactive B cells and plasma cells. By direct comparison of NGS-based clonality assessment with the conventional BIOMED-2/GeneScan analysis to detect IG gene rearrangements in undissected cHL-tumor tissue, our study demonstrated a significantly improved performance of the IG-NGS assay. The clonotypes detected by IG-NGS were shown to be of neoplastic cell origin, based on the identification of identical IG gene rearrangements in HRS-enriched cell fractions as compared to whole tissue specimens. Furthermore, our data revealed evidence for biclonality in two cHL cases, which was confirmed by single-cell analysis in one of the cases.
The recently developed IG-NGS clonality assay of the EuroClonality-NGS Working Group set out to improve clonality detection in lymphoid malignancies and has been validated for different mature B-cell lymphoma subtypes, but not yet for cHL24,26. Here, we investigated the applicability of the IG-NGS assay to detect clonal IG gene rearrangements in cHL. To define threshold values for tumor clone identification in cHL in the context of this study, clonality data of reactive lymphoproliferative samples without clear indications for EBV positivity were used to establish relative B-cell clone frequencies in non-malignant tissues. Identical clonotypes could be identified in duplicate analysis of the reactive lymphoproliferative samples with slightly increased frequencies compared to less-abundant background clonotypes.
On the basis of our defined thresholds, clonal IG gene rearrangements were detected with IG-NGS in 88% of the FF samples and 56% of the FFPE specimens, often at multiple targets for both IGH and IGK. Since all clonotypes were detected in duplicate in both FF and FFPE, these likely represent true clonal rearrangements and not artifacts. Furthermore, the identified dominant clonotypes showed no overlap between different samples, excluding cross-contamination or index-hopping. As expected, cHL tissues with relative higher frequencies of CD30-positive HRS cells as a proportion of total B cells showed more pronounced dominant clonotypes (Supplementary Fig. S11). Furthermore, a greater number of clonal IG gene rearrangements were detected in FF as compared to FFPE tissue, probably related to a higher genomic DNA integrity in FF material. However, each clonal rearrangement identified in FFPE was also detected in the paired FF cHL sample. In addition, IG-NGS detected clonality at more IG targets and additional cHL cases as compared to the BIOMED-2/EuroClonality assay, demonstrating the additive value of this approach. A recent study analyzed 120 non-Hodgkin lymphoma cases with IG-NGS26, where clonal IG gene rearrangements were detected in 96% of the lymphoma cases. This demonstrates that the identification of clonal IG gene rearrangements in cHL with NGS is less efficient (56% in FFPE, 88% in FF), most likely due to the lower fraction of malignant HRS cells within the tumor specimens. Studies in larger cohorts are required to firmly establish the clonality detection rate in cHL.
In three FF and seven FFPE samples, identical IG gene rearrangements were detected in top 3 duplicate clonotypes that did not reach our defined thresholds for clonality, and were indicated as ‘possible minor clones with unknown significance’ (Supplementary Table S4). In most of the FFPE samples, these clonotypes reached the thresholds for clonality in the corresponding FF sample and could therefore, in combination with the estimated tumor cell count, be considered as tumor-related clones for which detection was less efficient. Other minor clones, especially in the presence of a dominant tumor clone, most likely represented reactive B- or plasma cell clones, which in some cases could be related to the EBV status (Table 1). The increased sensitivity and specificity of IG-NGS relates to the smaller amplicons of the IG-targets in the assay in combination with a different primer design compared to the BIOMED-2/EuroClonality assay, the deep sequencing technique, and the information of specific clonotypes with the corresponding nucleotide sequences. Therefore, minor clones can be identified that otherwise would be blurred by the polyclonal background of conventional BIOMED-2/GeneScan analysis. On the other hand, all three frameworks for IGHV-IGHD-IGHJ rearrangements are analyzed in the BIOMED-2/EuroClonality assay, whereas the current IG-NGS assay is focussed on FR3.
Our studies demonstrate the presence of SHM in IGHV and to a lesser extent in IGKV gene regions of the malignant HRS cells (Supplementary Fig. S6), which is in line with previous findings8,9,35. Notably, 12 out of 14 cases displayed productive clonal IGHV-IGHD-IGHJ FR3 gene rearrangements, with CDR3 open reading frames ranging from 14 to 32 amino acids (Supplementary Tables S4 and S6). This is in concordance with a previous study, demonstrating that the majority of HRS IG gene rearrangements are functional9. However, HRS cells lack expression of the B-cell receptor (BCR), which is most likely due to the absence of active IG gene-specific transcription9 and epigenetic silencing of the BCR locus36. Nonetheless, IG gene rearrangements in cHL can serve as informative molecular fingerprints for clonality testing of cHL. To confirm that the observed dominant clonotypes in undissected tumor-tissue samples were derived from neoplastic cell origin, HRS cells were enriched by flow cytometry (FFPE tissue) and LMD (FF tissue). IG-NGS clonality testing showed identical clonal gene rearrangements and corresponding clonotypes at a higher abundancy in the isolated HRS-cell fraction as compared to whole-tumor tissue DNA, substantiating their neoplastic cell origin.
Furthermore, our data revealed the presence of multiple clonal rearrangements, which suggests the occurrence of biclonality in cHL (Supplementary Fig. S8). Although true bi- or oligoclonality is an infrequent phenomenon in B-cell lymphoproliferative disorders37,38, biclonality has been reported mostly in the context of composite lymphomas39,40,41, and also within single lymphoma subtypes42,43 and in immunosuppressed patients44,45. It is known that, especially in patients with a history of infectious mononucleosis or immune deficiencies, EBV latent antigens can regulate or mimic different survival and proliferative signaling pathways, and thereby promote lymphomagenesis46,47,48. Since both of our cases with suspected biclonality showed EBV positivity, this might contribute to clonal diversity with the detection of two clonal IGHD-IGHJ and one IGHV-IGHD-IGHJ rearrangement in case 8, and multiple IGK gene rearrangements in case 9, which do not fit within a single B-cell clone (Supplementary Fig. S8). Besides this immunogenetic evidence for the involvement of at least two clones, we also employed single-cell CNV analysis on DEPArray digital sorted HRS cells of case 9, which revealed two clusters of distinct CNV profiles. Each of the two HRS clusters displayed prominent intraclonal heterogeneity, which is likely related to the genetic instability of HRS cells33,34, and is in concordance with a recent study of Mangano et al.31. Unfortunately, such single-cell analysis could not be performed on case 8, due to the inferior FFPE-DNA quality.
In conclusion, IG-NGS clonality testing is a more accurate and sensitive assay for clonality detection in whole-tumor tissue DNA of cHL samples compared to the conventional BIOMED-2/EuroClonality assay. Additionally, this new IG-NGS based clonality testing will be an ideal tool in clinical diagnostics for recurrent disease, since clonal comparison based on nucleotide sequences and assigned clonotypes is more reliable than comparing fragment lengths, as has been shown in non-Hodgkin lymphoma patients24,49. Moreover, this approach allows more detailed analysis on the clonal composition and its diversity that underlies cHL pathogenesis.
Data availability
All next-generation sequencing datafiles related to clonality assessment presented in this study are available from the corresponding author on reasonable request.
Change history
15 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41379-021-00986-5
References
Küppers, R. The biology of Hodgkin’s lymphoma. Nat. Rev. Cancer 9, 15–27 (2009).
Weniger, M. A. & Küppers, R. Molecular biology of Hodgkin lymphoma. Leukemia 35, 968–981 (2021).
Schmitz, R., Stanelle, J., Hansmann, M. L. & Küppers, R. Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annu. Rev. Pathol. 4, 151–174 (2009).
Kuppers, R. et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 91, 10962–10966 (1994).
Tamaru, J., Hummel, M., Zemlin, M., Kalvelage, B. & Stein, H. Hodgkin’s disease with a B-cell phenotype often shows a VDJ rearrangement and somatic mutations in the VH genes. Blood 84, 708–715 (1994).
Deng, F., Lü, G., Li, G. & Yang, G. Hodgkin’s disease: immunoglobulin heavy and light chain gene rearrangements revealed in single Hodgkin/Reed-Sternberg cells. Mol. Pathol. 52, 37–41 (1999).
Tapia, G., Sanz, C., Mate, J. L., Munoz-Marmol, A. M. & Ariza, A. Improved clonality detection in Hodgkin lymphoma using the BIOMED-2-based heavy and kappa chain assay: a paraffin-embedded tissue study. Histopathology 60, 768–773 (2012).
Kanzler, H., Kuppers, R., Hansmann, M. L. & Rajewsky, K. Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 184, 1495–1505 (1996).
Marafioti, T. et al. Hodgkin and reed-sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood 95, 1443–1450 (2000).
Brauninger, A., Wacker, H. H., Rajewsky, K., Kuppers, R. & Hansmann, M. L. Typing the histogenetic origin of the tumor cells of lymphocyte-rich classical Hodgkin’s lymphoma in relation to tumor cells of classical and lymphocyte-predominance Hodgkin’s lymphoma. Cancer Res. 63, 1644–1651 (2003).
Fend, F. Classical Hodgkin lymphoma and its differential diagnoses. Diagn. Histopathol. 21, 400–407 (2015).
Parente, P., Zanelli, M., Sanguedolce, F., Mastracci, L. & Graziano, P. Hodgkin Reed-Sternberg-like cells in non-Hodgkin lymphoma. Diagnostics (Basel) 10, 1019 (2020).
van Dongen, J. J. et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17, 2257–2317 (2003).
van Krieken, J. H. et al. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 21, 201–206 (2007).
Langerak, A. W. et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia 26, 2159–2171 (2012).
Evans, P. A. et al. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 21, 207–214 (2007).
Chute, D. J. et al. Detection of immunoglobulin heavy chain gene rearrangements in classic hodgkin lymphoma using commercially available BIOMED-2 primers. Diagn. Mol. Pathol. 17, 65–72 (2008).
Hebeda, K. M., Van Altena, M. C., Rombout, P., Van Krieken, J. H. & Groenen, P. J. PCR clonality detection in Hodgkin lymphoma. J. Hematop. 2, 34–41 (2009).
Burack, W. R., Laughlin, T. S., Friedberg, J. W., Spence, J. M. & Rothberg, P. G. PCR assays detect B-lymphocyte clonality in formalin-fixed, paraffin-embedded specimens of classical hodgkin lymphoma without microdissection. Am. J. Clin. Pathol. 134, 104–111 (2010).
Hartmann, S., Helling, A., Doring, C., Renne, C. & Hansmann, M. L. Clonality testing of malignant lymphomas with the BIOMED-2 primers in a large cohort of 1969 primary and consultant biopsies. Pathol. Res. Pract. 209, 495–502 (2013).
Ghorbian, S. et al. Molecular analysis of IGH and incomplete IGH D-J clonality gene rearrangements in Hodgkin lymphoma malignancies. Clin. Lab. 61, 951–955 (2015).
Ghorbian, S., Jahanzad, I., Javadi, G. R. & Sakhinia, E. Evaluation of IGK and IGL molecular gene rearrangements according to the BIOMED-2 protocols for clinical diagnosis of Hodgkin lymphoma. Hematology 21, 133–137 (2016).
Han, S. et al. Improved clonality detection in Hodgkin lymphoma using a semi-nested modification of the BIOMED-2 PCR assay for IGH and IGK rearrangements: a paraffin-embedded tissue study. Pathol. Int. 68, 287–293 (2018).
Scheijen, B. et al. Next-generation sequencing of immunoglobulin gene rearrangements for clonality assessment: a technical feasibility study by EuroClonality-NGS. Leukemia 33, 2227–2240 (2019).
Swerdlow, S. H., Campo, E., Harris, N. L. & Pileri, S. A. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (International Agency for Research on Cancer, 2017).
van den Brand, M. et al. Next-generation sequencing-based clonality assessment of Ig gene rearrangements: a multicenter validation study by EuroClonality-NGS. J. Mol. Diagn. 23, 1105–1115 (2021).
Bystry, V. et al. ARResT/Interrogate: an interactive immunoprofiler for IG/TR NGS data. Bioinformatics 33, 435–437 (2017).
Knecht, H. et al. Quality control and quantification in IG/TR next-generation sequencing marker identification: protocols and bioinformatic functionalities by EuroClonality-NGS. Leukemia 33, 2254–2265 (2019).
Juskevicius, D. et al. Novel cell enrichment technique for robust genetic analysis of archival classical Hodgkin lymphoma tissues. Lab. Invest. 98, 1487–1499 (2018).
Leenders, A. M. et al. Multiple immunoglobulin kappa gene rearrangements within a single clone unraveled by next-generation sequencing-based clonality assessment. J. Mol. Diagn. 23, 1097–1104 (2021).
Mangano, C. et al. Precise detection of genomic imbalances at single-cell resolution reveals intra-patient heterogeneity in Hodgkin’s lymphoma. Blood Cancer J. 9, 92 (2019).
Bolognesi, C. et al. Digital sorting of pure cell populations enables unambiguous genetic analysis of heterogeneous formalin-fixed paraffin-embedded tumors by next generation sequencing. Sci. Rep. 6, 20944 (2016).
Cuceu, C., et al. Chromosomal instability in Hodgkin lymphoma: an in-depth review and perspectives. Cancers (Basel) 10, 91 (2018).
Re, D., Zander, T., Diehl, V. & Wolf, J. Genetic instability in Hodgkin’s lymphoma. Ann. Oncol. 13(Suppl. 1), 19–22 (2002).
Vockerodt, M. et al. Detection of clonal Hodgkin and Reed-Sternberg cells with identical somatically mutated and rearranged VH genes in different biopsies in relapsed Hodgkin’s disease. Blood 92, 2899–2907 (1998).
Ushmorov, A. et al. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood 104, 3326–3334 (2004).
Sanchez, M. L. et al. Incidence and clinicobiologic characteristics of leukemic B-cell chronic lymphoproliferative disorders with more than one B-cell clone. Blood 102, 2994–3002 (2003).
Hengeveld, P. J., Levin, M. D., Kolijn, P. M. & Langerak, A. W. Reading the B-cell receptor immunome in chronic lymphocytic leukemia: revelations and applications. Exp. Hematol. 93, 14–24 (2021).
Boiocchi, L. et al. Composite chronic lymphocytic leukemia/small lymphocytic lymphoma and follicular lymphoma are biclonal lymphomas: a report of two cases. Am. J. Clin. Pathol. 137, 647–659 (2012).
Fend, F. et al. Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas. A molecular analysis using laser capture microdissection. Am. J. Pathol. 154, 1857–1866 (1999).
Cabras, A. D. et al. Biclonality of gastric lymphomas. Lab. Invest. 81, 961–967 (2001).
Delville, J. P. et al. Biclonal low grade B-cell lymphoma confirmed by both flow cytometry and karyotypic analysis, in spite of a normal kappa/lambda Ig light chain ratio. Am. J. Hematol. 82, 473–480 (2007).
Edinger, J. T., Lorenzo, C. R., Breneman, D. L. & Swerdlow, S. H. Primary cutaneous marginal zone lymphoma with subclinical cutaneous involvement and biclonality. J. Cutan. Pathol. 38, 724–730 (2011).
Cleary, M. L. & Sklar, J. Lymphoproliferative disorders in cardiac transplant recipients are multiclonal lymphomas. Lancet 2, 489–493 (1984).
Foo, W. C. et al. Concurrent classical Hodgkin lymphoma and plasmablastic lymphoma in a patient with chronic lymphocytic leukemia/small lymphocytic lymphoma treated with fludarabine: a dimorphic presentation of iatrogenic immunodeficiency-associated lymphoproliferative disorder with evidence suggestive of multiclonal transformability of B cells by Epstein-Barr virus. Hum. Pathol. 41, 1802–1808 (2010).
Saha, A. & Robertson, E. S. Mechanisms of B-Cell oncogenesis induced by Epstein-Barr virus. J. Virol. 93, e00238-19 (2019).
Murray, P. G. & Young, L. S. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood 134, 591–596 (2019).
Shannon-Lowe, C., Rickinson, A. B. & Bell, A. I. Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372 20160271 (2017).
Lee, S. E. et al. Clonal relationships in recurrent B-cell lymphomas. Oncotarget 7, 12359–12371 (2016).
Acknowledgements
The authors would like to thank everyone who contributed to the IG-NGS Biological Validation study26, and the EuroClonality-NGS Working Group, coordinated by Ton Langerak, for the IG-NGS clonality data of the reactive lymphoproliferative samples; Rob Woestenenk for help with flow cytometric isolation of the HRS cells; technicians from the department of Human Genetics for preparing the next-generation sequence runs, and Menarini for performing DEPArray™ single cell isolation and genomic profiling. Illustrations were created with BioRender.com.
Funding
This project was funded by the Dutch Cancer Society (KWF-11137) and the Dutch Health Insurers’ Innovation Fund (Project no. 17-179).
Author information
Authors and Affiliations
Contributions
H.v.K. and B.S. designed the research and conceived the project. D.v.B., J.R., D.H., and J.L. performed the experimental research, D.v.B., M.vd.B., K.H., P.G., and B.S. analyzed the data, S.P.N. was responsible for the bioinformatic CNV data analysis. D.v.B. and B.S. wrote the manuscript. M.vd.B., K.H., and H.v.K. examined the histopathology and together with P.G. reviewed the manuscript. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Ethics Approval
All experiments with human tissues were performed in accordance with the Declaration of Helsinki. This study received approval of the institutional CMO review board (Approval No. 2020-6390).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Due to error in table 3.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Bladel, D.A.G., van den Brand, M., Rijntjes, J. et al. Clonality assessment and detection of clonal diversity in classic Hodgkin lymphoma by next-generation sequencing of immunoglobulin gene rearrangements. Mod Pathol 35, 757–766 (2022). https://doi.org/10.1038/s41379-021-00983-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00983-8
This article is cited by
-
Diagnostic and prognostic molecular pathology of lymphoid malignancies
Virchows Archiv (2023)