Abstract
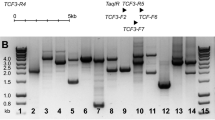
Although reciprocal chromosomal translocations are not typical for B-cell chronic lymphocytic leukemia (B-CLL), we identified the novel t(1;6)(p35.3;p25.2) in eight patients with this disorder. Interestingly, all cases showed lack of somatically mutated IgVH. Clinical, morphological, immunologic, and genetic features of these patients are described. Briefly, the age ranged from 33 to 81 years (median: 62.5 years) and the sex ratio was 6M:2F. Most of the patients (6/8) presented with advanced clinical stage. Therapy was required in seven cases. After a median follow-up of 28 months, five patients are alive and three died from disease evolution. Three cases developed transformation into diffuse large B-cell lymphoma. Translocation t(1;6) was found as the primary karyotypic abnormality in three patients. Additional chromosomal aberrations included changes frequently found in unmutated B-CLL, that is, del(11)(q), trisomy 12 and 17p aberrations. Fluorescence in situ hybridization analysis performed in seven cases allowed us to map the t(1;6) breakpoints to the 1p35.3 and 6p25.2 chromosomal bands, respectively. The latter breakpoint was located in the genomic region coding for MUM1/IRF4, one of the key regulators of lymphocyte development and proliferation, suggesting involvement of this gene in the t(1;6). Molecular characterization of the t(1;6)(p35.3;p25.2), exclusively found in unmutated subtype of B-CLL, is in progress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS . Clinical staging of chronic lymphocytic leukemia. Blood 1975; 46: 219–234.
Binet JL, Auquier A, Dighiero G, Chastang C, Piguet H, Goasguen J et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer 1981; 48: 198–206.
Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood 2002; 100: 1177–1184.
Vasconcelos Y, Davi F, Levy V, Oppezzo P, Magnac C, Michel A et al. Binet's staging system and VH genes are independent but complementary prognostic indicators in chronic lymphocytic leukemia. J Clin Oncol 2003; 21: 3928–3932.
Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–1847.
Guarini A, Gaidano G, Mauro FR, Capello D, Mancini F, De Propris MS et al. Chronic lymphocytic leukemia patients with highly stable and indolent disease show distinctive phenotypic and genotypic features. Blood 2003; 102: 1035–1041.
Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK . Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–1854.
Kröber A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood 2002; 100: 1410–1416.
Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med 2001; 194: 1625–1638.
Döhner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916.
Geisler CH, Philip P, Christensen BE, Hou-Jensen K, Pedersen NT, Jensen OM et al. In B-cell chronic lymphocytic leukaemia chromosome 17 abnormalities and not trisomy 12 are the single most important cytogenetic abnormalities for the prognosis: a cytogenetic and immunophenotypic study of 480 unselected newly diagnosed patients. Leukemia Res 1997; 21: 1011–1023.
Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med 1990; 323: 720–724.
Chevallier P, Penther D, Avet-Loiseau H, Robillard N, Ifrah N, Mahe B et al. CD38 expression and secondary 17p deletion are important prognostic factors in chronic lymphocytic leukaemia. Br J Haematol 2002; 116: 142–150.
Willis TG, Dyer MJ . The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood 2000; 96: 808–822.
Lin K, Sherrington PD, Dennis M, Matrai Z, Cawley JC, Pettitt AR . Relationship between p53 dysfunction, CD38 expression, and IgV(H) mutation in chronic lymphocytic leukemia. Blood 2002; 100: 1404–1409.
Mitelman F (ed). ISCN (1995) Guidelines for Cancer Cytogenetics. Supplement to An International System for Human Cytogenetic Nomenclature. Basel: Karger, 1995.
Dierlamm J, Wlodarska I, Michaux L, La-Starza R, Zeller W, Mecucci C et al. Successful use of the same slide for consecutive fluorescence in situ hybridization experiments. Genes Chromosomes Cancer 1996; 16: 261–264.
Martin-Subero JI, Harder L, Gesk S, Schlegelberger B, Grote W, Martinez-Climent JA et al. Interphase FISH assays for the detection of translocations with breakpoints in immunoglobulin light chain loci. Int J Cancer 2002; 98: 470–474.
Aubin J, Davi F, Nguyen-Salomon F, Leboeuf D, Debert C, Taher M et al. Description of a novel FR1 IgH PCR strategy and its comparison with three other strategies for the detection of clonality in B cell malignancies. Leukemia 1995; 9: 471–479.
Langerak AW, Szczepanski T, van der BM, Wolvers-Tettero IL, van Dongen JJ . Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997; 11: 2192–2199.
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR et al. Proposals for the classification of chronic (mature) B and T lymphoid leukaemias. French–American–British (FAB) Cooperative Group. J Clin Pathol 1989; 42: 567–584.
Criel A, Michaux L, Wolf-Peeters C . The concept of typical and atypical chronic lymphocytic leukaemia. Leukemia Lymphoma 1999; 33: 33–45.
Matutes E, Owusu-Ankomah K, Morilla R, Garcia MJ, Houlihan A, Que TH et al. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994; 8: 1640–1645.
Knight SJ, Flint J . Perfect endings: a review of subtelomeric probes and their use in clinical diagnosis. J Med Genet 2000; 37: 401–409.
Timar B, Fulop Z, Csernus B, Angster C, Bognar A, Szepesi A et al. Relationship between the mutational status of VH genes and pathogenesis of diffuse large B-cell lymphoma in Richter's syndrome. Leukemia 2004; 18: 326–330.
Oscier DG, Thompsett A, Zhu D, Stevenson FK . Differential rates of somatic hypermutation in V(H) genes among subsets of chronic lymphocytic leukemia defined by chromosomal abnormalities. Blood 1997; 89: 4153–4160.
Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 1997; 275: 540–543.
Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B et al. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 1997; 17: 226–230.
Acknowledgements
This work presents research results of the Belgian program of Interuniversity Poles of attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The scientific responsibility is assumed by the authors. The study was partially supported by the Fund for Scientific Research of Flanders (FWO - Vlaanderen), Grant no. G.0338.01.
We thank U. Pluys for excellent technical assistance, P. Vannuffel for performing IgVH analyses, V.Deneys for performing immunophenotypic analyses, and R. Logist for secretarial assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Rights and permissions
About this article
Cite this article
Michaux, L., Wlodarska, I., Rack, K. et al. Translocation t(1;6)(p35.3;p25.2): a new recurrent aberration in ‘unmutated’ B-CLL. Leukemia 19, 77–82 (2005). https://doi.org/10.1038/sj.leu.2403543
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2403543
Keywords
This article is cited by
-
A Japanese case of chronic lymphocytic leukemia with t (1;6)
Experimental Hematology & Oncology (2012)
-
Germline copy number variation associated with Mendelian inheritance of CLL in two families
Leukemia (2012)
-
IRF4 Gene Rearrangements Define a Subgroup of CD30-Positive Cutaneous T-Cell Lymphoma: A Study of 54 Cases
Journal of Investigative Dermatology (2010)
-
Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas
Leukemia (2009)