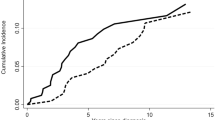
Abstract
The majority of patients with acute promyelocytic leukemia (APL) possess either a bcr1 or a bcr3 type fusion between PML and RARα genes. The junction sequences may possibly be a target for immune response and influence susceptibility to the disease. In this case, HLA class I allele frequencies would be different between bcr1 and bcr3 patients. To test this hypothesis, we typed 102 APL patients for HLA-A, -B and -Cw alleles. The A*1, A*30, B*51, B*41, Cw*0602, and Cw*1701 alleles showed a different distribution between bcr1 and bcr3 patients, but in no case was this statistically significant after correction for the number of comparisons or was confirmed in an independent panel. Moreover, no difference was detected between bcr1 and bcr3 when HLA alleles were grouped according to their peptide binding specificities. Comparing HLA frequencies, clinical features at diagnosis and clinical outcome of the 64 patients homogeneously treated with all-trans retinoic acid and idarubicin (AIDA protocol) we observed a statistically significant association between HLA-B*13 and risk of relapse by univariate and multivariate regression analysis. Should this finding be confirmed in larger future studies, this observation would be of outmost importance in identifying patients at high risk of relapse in which more aggressive consolidation therapies should be used.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Hester JP, Rossen R, Trujillo J, McCredie KB, Freireich EJ . Frequency of HLA antigens in chronic myelocitic leukemia S Med J 1977 70: 691–693
Navarrete C, Alonso A, Awad J, McCloskey D, Ganesan TS, Amess J, Lister TA, Festenstein H . HLA class I and class II antigens association in acute leukemias J Immunogenet 1986 13: 77–84
Annino L, Ferrari A, Laurenti L, Perrone MP, Romani C, Pacchiarotti A, Girelli G, Mandelli F . HLA typing in hairy cell leukemia Leuk Lymphoma 1994 14: 63–65
Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P . PMLRAR homodimers: distinct DNA properties and heterodimeric interactions with RXR EMBO J 1993 12: 3171–3182
Pandolfi PP, Alcalay M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F, Rochette-Egly C, Gaub MP, Chambon P, Pelicci PG . Genomic variability and alternative splicings generate multiple PML/RARα transcripts that encode aberrant PML proteins and PML/RARα isoforms in acute promyelocytic leukemias EMBO J 1992 11: 1397–1407
Bocchia M, Korontsvit T, Xu Q, Mackinnon S, Yang SY, Sette A, Scheinberg DA . Specific human cellular immunity to bcr-abl oncogene-derived peptides Blood 1996 87: 3587–3592
Greco G, Fruci D, Accapezzato D, Barnaba V, Nisini R, Alimena G, Montefusco E, Vigneti E, Butler R, Tanigaki N, Tosi R . Two bcr-abl junction peptides bind HLA-A3 molecules and allow specific induction of human cytotoxic T lymphocytes Leukemia 1996 10: 639–699
Lecchi M, Lovisone E, Genetta C, Peruccio D, Resegotti L, Richiardi P . Gamma-IFN induces a differential expression of HLA-DR, DQ and DP antigens on peripheral blood myeloid leukemic blasts at various stages of differentiation Leukemia Res 1989 13: 221–226
Jinnai I . In vitro growth response to G-CSF and GM-CSF by bone marrow cells of patients with acute myeloyd leukemia Leukemia Res 1990 14: 227–240
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DAG, Gralnick HR, Sultan C . Proposal for the classification of the acute leukemias Br J Haematol 1976 33: 451–458
Mandelli F, Diverio D, Avvisati G, Luciano A, Barbui T, Bernasconi C, Broccia G, Cerri R, Falda M, Fioritoni G, Leoni F, Liso V, Petti MC, Rodeghiero F, Saglio G, Vegna ML, Visani G, Jehn U, Villemze R, Muus P, Pelicci PG, Biondi A, Lo Coco F . Molecular remission in the PML/RARa positive acute promyelocytic leukemia by combined all-trans retinoic acid and idarubicin AIDA therapy Blood 1997 90: 1014–1021
Fasano ME, Praticò L, Brancatello F, Mazzola G, Cravero T, Bersanti M, Curtoni ES . Caucasian Italian normal. In: Terasaki P, Gjertson D (eds) HLA 1998 American Society for Histocompatibility and Immunogenetics: Lenexa, KS 1999 p 162
Francia di Celle P, Gay E, Marin F, Maderno P, Berrino M, Dall'Omo A, Curtoni ES . Caucasian Italian normal. In: Terasaki P, Gjertson D (eds) HLA 1998 American Society for Histocompatibility and Immunogenetics: Lenexa, KS 1999 p 163
Rendine S, Borelli I, Barbanti M, Sacchi N, Roggero S, Curtoni ES . HLA polymorphisms in Italian bone marrow donors: a regional analysis Tissue Antigens 1998 52: 135–146
Clayton J, Lonjou C . Allele and haplotype frequencies for HLA loci in various ethnic groups. In: D Charron (ed) Genetic Diversity of HLA. Functional and Medical Implication EDK: Paris 1997 1: p 675
Diverio D, Lo Coco F, D'Adamo F, Biondi A, Fagioli M, Grignani F, Rambaldi A, Rossi V, Avvisati G, Petti MC, Testi AM, Liso V, Specchia G, Fioritoni G, Recchia A, Frassoni F, Ciolli S, Pelicci PG for the Italian Cooperative Study group GIMEMA . Identification of DNA rearrangements at the retinoic acid receptor alfa RARa locus in all patients with acute promyelocytic leukemia APL and mapping of APL breakpoints within the RARa second intron Blood 1992 79: 3331–3336
Diverio D, Riccioni R, Pistilli A, Buffolino S, Avvisati G, Mandelli F, Lo Coco F . Improved rapid detection of the PML/RARa fusion gene in acute promyelocytic leukemia Leukemia 1996 10: 1214–1216
Chomczynsky P, Sacchi N . Single step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction Anal Biochem 1987 162: 156–158
Tonks S, Marsh SGE, Bunce M, Moses JH, Krausa P, Sadler AM, Petronzelli F, Bodmer JG . HLA class I DNA typing study. In: D Charron (ed). Genetic Diversity of HLA. Functional and Medical Implication EDK: Paris 1997; 1: pp188–193
Oh S, Fleischhauer K, Yang SY . Isoelectric focusing subtypes of HLA-A can be defined by oligonucleotide typing Tissue Antigens 1993 41: 135–142
Fleischhauer K, Zino E, Bordignon C, Benazzi E . Complete, generic and extensive fine-specificity typing of the HLA-B locus by the PCR-SSOP method Tissue Antigens 1995 46: 281–292
Zino E, Severini GM, Mazzi B, Bordignon C, Benazzi E, Fleischhauer K . Sequencing of a new HLA-A*32 subtype A*3202 Immunogenetics 1996 45: 76–77
Grundschober C, Rufer N, Sanchez-Mazas A, Madrigal A, Jeannet M, Roosnek E, Tiercy JM . Molecular characterization of HLA-C incompatibilities in HLA-ABDR-matched unrelated bone marrow donor-recipient pairs. Sequence of two new Cw alleles Cw*02023 and Cw*0707 and recognition by cytotoxic T lymphocytes Tissue Antigens 1997 49: 612–623
Sidney J, Grey HM, Kubo RT, Sette A . Practical, biochemical and evolutionary implications of the discovery of HLA class supermotifs Immunol Today 1996 17: 261–266
Gambacorti-Passerini C, Bertazzoli C, Dermime S, Scardino A, Schendel D, Parmiani G . Mapping of HLA class I binding motifs in 44 fusion proteins involved in human cancers Clin Cancer Res 1997 3: 675–683
Bocchia M, Wentworth PA, Southwood S, Sidney J, McGraw K, Scheinberg DA, Sette A . Specific binding of leukemia oncogene fusion protein peptides to HLA class I molecules Blood 1995 85: 2680–2684
Bolognesi E, D'Alfonso S, Greco G, Tanigaki N, Fleischhauer C, Cimino G, Rapanotti C, Diverio D, Momigliano-Richiardi P . HLA class I and susceptibility to acute promyelocytic leukemia Eur J Immunogenet 1998 25: S56
Zheng P, Guo Y, Niu Q, Levy DE, Dyck JA, Lu S, Sheiman LA, Liu Y . Proto-oncogene PML controls genes devoted to MHC class I antigen presentation Nature 1998 396: 373–376
Acknowledgements
This work was supported by grants from FIRC, the Italian Foundation for Cancer Research, and from Associazione Italiana per la Leucemia, sezione di Roma (ROMAIL). E Bolognesi is a recipient of a fellowship from Gruppo di Cooperazione in Cancerologia. MC Rapanotti is a recipient of a fellowship from FIRC. We are grateful to Dr R Tosi for critically reviewing the manuscript.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bolognesi, E., Cimino, G., Diverio, D. et al. HLA class I in acute promyelocytic leukemia (APL): possible correlation with clinical outcome. Leukemia 14, 393–398 (2000). https://doi.org/10.1038/sj.leu.2401691
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2401691
Keywords
This article is cited by
-
Incidence of extramedullary disease in patients with acute promyelocytic leukemia: a single-institution experience
International Journal of Hematology (2009)
-
Management of acute promyelocytic leukemia
Current Oncology Reports (2002)
-
Is there any genetic susceptibility to acute promyelocytic leukemia?
Leukemia (2000)