Abstract
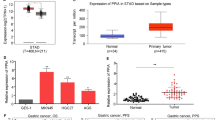
Multiple genetic alterations are attributed to gastric cancer (GC); however, only a few critical genes have been identified so far. In this study, we isolated and characterized a novel gene p42.3, represented as tumor-specific and mitosis phase-dependent expression protein in GC cell line BGC823. Our data showed that the expression of p42.3 was cell cycle-dependent in GC cell lines. Moreover, p42.3 was specifically expressed in primary GC tissues but not in the matched normal mucosa of stomach, and this gene was expressed in diverse embryonic tissues. Furthermore, significant suppression of cell proliferation and tumorigenicity were detected and G2/M phase arrest was observed in cell line BGC823 depleted of p42.3 expression by RNAi technique, and we confirmed the expression changes of cyclin B1 and Chk2 following the silence of p42.3. Taken together, we cloned and characterized p42.3 gene that was specifically expressed in GC tumors but not in normal gastric mucosa, and the gene was associated with M-phase regulation. Moreover, p42.3 might be involved in cell proliferation and tumorigenesis; therefore, this gene might have potential applications in the diagnosis or treatment of GC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Chaturvedi P, Eng WK, Zhu Y, Mattern MR, Mishra R, Hurle MR et al. (1999). Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene 18: 4047–4054.
Cobb JA, Bjergbaek L . (2006). RecQ helicases: lessons from model organisms. Nucleic Acids Res 34: 4106–4114.
Deng CX . (2006). BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 34: 1416–1426.
Deng H, Guo RF, Li WM, Zhao M, Lu YY . (2005). Matrix metalloproteinase 11 depletion inhibits cell proliferation in gastric cancer cells. Biochem Biophys Res Commun 326: 274–281.
Elledge SJ . (1996). Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1673.
Hanahan D, Weinberg RA . (2000). The hallmarks of cancer. Cell 100: 57–70.
Hartwell LH, Kastan MB . (1994). Cell cycle control and cancer. Science 266: 1821–1828.
Herness EA, Naz RK . (2003). A novel human prostate-specific gene-1 (HPG-1): molecular cloning, sequencing, and its potential involvement in prostate Carcinogenesis. Cancer Res 63: 329–336.
Ji JF, Chen X, Leung SY, Chi JT, Chu KM, Siu TY et al. (2002). Comprehensive analysis of the gene expression profiles in human gastric cancer cell lines. Oncogene 21: 6549–6556.
Kay BK, Williamson MP, Sudol M . (2000). The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14: 231–241.
Kumagai A, Dunphy WG . (1999). Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev 13: 1067–1072.
Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ . (2002). Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124.
Lu M, Zhang LP, Maul RS, Sartippour MR, Norris A, Whitelegge J et al. (2005). The novel gene EG-1 stimulates cellular proliferation. Cancer Res 65: 6159–6166.
Murata T, Furushima K, Hirano M, Kiyonari H, Nakamura M, Suda Y . (2004). Ang is a novel gene expressed in early neuroectoderm, but its null mutant exhibits no obvious phenotype. Gene Expr Patterns 5: 171–178.
Murray AW . (2004). Recycling the cell cycle: cyclins revisited. Cell 116: 221–234.
Nasmyth K . (1996). Viewpoint: putting the cell cycle in order. Science 274: 1643–1645.
Pinto-Santini D, Salama NR . (2005). The biology of helicobacter pylori infection, a major risk factor for gastric adenocarcinoma. Cancer Epidemiol Biomarkers Prev 14: 1853–1858.
Santamaria-Kisiel L, Shaw GS, Shaw GS . (2006). Calcium-dependent and -independent interactions of the S100 protein family. Biochem J 396: 201–214.
Stock M, Otto F . (2005). Gene deregulation in gastric cancer. Gene 360: 1–19.
Strauberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS . (2002). Generation and initial analysis of more than 15 000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99: 16899–16903.
Tang ZB, Zhao M, Ji JF, Yang GB, Hu FL, He JS et al. (2004). Overexpression of gastrin and c-met protein involved in human gastric carcinomas and intestinal metaplasia. Oncol Rep 11: 333–339.
Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K et al. (1999). Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res 59: 5438–5442.
You WC, Li JY, Zhang L, Jin ML, Chang YS, Ma JL et al. (2005). Etiology and prevention of gastric cancer: a population study in a high risk area of China. Chin J Diq Dis 6: 149–154.
Zeng JZ, Wang HY, Chen ZJ, Ulrich A, Wu MC et al. (2002). Molecular cloning and characterization of a novel gene which is highly expressed in hepatocellular carcinoma. Oncogene 21: 4932–4943.
Zhang PN, McAlinden A, Li S, Schumacher T, Wang HL, Hu S et al. (2001). The amino-terminal heptad repeats of the coiled-coil neck domain of pulmonary surfactant protein D are necessary for the assembly of trimeric subunits and dodecamers. J Biol Chem 276: 19862–19870.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (39625016), State Key Basic Research Program of China (2004CB518708) and the National High Technology Research and Development Program of China (2002BA711A11).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary information
Rights and permissions
About this article
Cite this article
Xu, X., Li, W., Fan, X. et al. Identification and characterization of a novel p42.3 gene as tumor-specific and mitosis phase-dependent expression in gastric cancer. Oncogene 26, 7371–7379 (2007). https://doi.org/10.1038/sj.onc.1210538
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210538
Keywords
This article is cited by
-
SAPCD2 promotes neuroblastoma progression by altering the subcellular distribution of E2F7
Cell Death & Disease (2022)
-
Overexpression of SAPCD2 correlates with proliferation and invasion of colorectal carcinoma cells
Cancer Cell International (2020)
-
C9orf140, a novel Axin1-interacting protein, mediates the negative feedback loop of Wnt/β-catenin signaling
Oncogene (2018)
-
Depletion of p42.3 gene inhibits proliferation and invasion in melanoma cells
Journal of Cancer Research and Clinical Oncology (2017)
-
Overexpression of p42.3 promotes cell proliferation, migration, and invasion in human gastric cancer cells
Tumor Biology (2016)