Abstract
A long-acting, thermostable bacterial cocaine esterase (CocE) has been identified that rapidly degrades cocaine with a KM of 1.33+0.085 μM. In vivo evaluation of CocE has shown protection against convulsant and lethal effects of cocaine in rodents, confirming the therapeutic potential of CocE against cocaine overdose. However, the current study is the first to evaluate the effects of CocE on cocaine brain levels. Positron emission tomogrpahy neuroimaging of [11C]cocaine was used to evaluate the time course of cocaine elimination from brain in the presence and absence of CocE in nonhuman primates. Systemic administration of CocE eliminated cocaine from the rhesus-monkey brain approximately three times faster than control conditions via peripheral actions through attenuating the input function from blood plasma. The efficiency of this process is sufficient to alleviate or prevent adverse central nervous system effects induced by cocaine. Although the present study used tracer doses of cocaine to access brain clearance, these findings further support the development of CocE for the treatment of acute cocaine toxicity.
Similar content being viewed by others
Introduction
Searching for sensitive methods to detect illicit cocaine, a group in Cambridge, England investigated the presence of cocaine biosensors in a Rhodococcus bacterium that inhabits the soil of the coca plant rhizosphere. They isolated, identified, sequenced and cloned a gene that encodes cocaine esterase (CocE), by this bacterium and degrades cocaine into inactive metabolites, ecgonine methyl ester and benzoic acid, with a KM of 1.33+0.085 μM.1 The possibility that such an enzyme could be useful in the treatment of cocaine overdose was noted by a group at the Scripps Research Institute in 2002.2,3 This group crystallized and further characterized the enzyme, determining the kinetics of CocE as: kcat=7.8 s−1 and KM=640 nM. In vivo evaluation of CocE’s actions against cocaine demonstrated the ability of the enzyme to protect against the convulsant and lethal effects of cocaine in rodents.4,5 Hence, the therapeutic potential of CocE against cocaine overdose has been confirmed.
The native CocE protein has a short enzymatic half-life in plasma at normal physiological temperature (37 °C). However, two amino acid changes based off in silico analysis were able to increase its half-life from 15 min to over 4 h without altering the enzyme’s hydrolytic activity.6 The stabilized form of the enzyme eliminates the cardiovascular effects of systemically delivered cocaine.7 Even if cocaine had initiated a seizure as indicated by EEG recordings, it could be quickly terminated by intravenous delivery of the enzyme.8 These data raised the interesting question regarding the site of CocE’s action. Due to its large size, the enzyme is unlikely to enter the brain following systemic administration. Accordingly, its effects are likely to be exclusively due to its capacity to act as a peripheral ‘sink’ to draw cocaine out of the brain and hydrolyze it in the periphery.
To confirm this theoretical framework, we prepared the double mutant CocE in a bacterial cell line that expressed no endotoxin, radiolabeled cocaine with [11C] and used positron emission tomogrpahy (PET) neuroimaging to evaluate the time course of cocaine elimination from brain in the presence and absence of CocE in rhesus monkeys. We hypothesized that the presence of CocE in plasma would accelerate the elimination of cocaine from brain. As a negative control, we evaluated the cocaine analog RTI-150 which lacks the benzoyl ester linkage of cocaine but has characteristic cocaine-like effects on behavior and dopamine neurochemistry in nonhuman primates.9, 10, 11 If our suppositions were correct, CocE should have no effect on the pharmacokinetic profile of RTI-150.
Materials and methods
Subjects
Three adult rhesus monkeys (RGg-9, RZq-8 and RLa-10) weighing between 8.1–9.8 kg served as subjects. Each subject was housed individually and fed Purina monkey chow (Ralston Purina, St Louis, MO, USA), fruits and vegetables. Water was continuously available. Animal care procedures strictly followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Emory University. All subjects received [11C]cocaine and two subjects (RGg-9 and RZq-8) also received [11C]RTI-150.
Radioligands
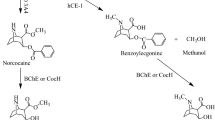
Cocaine was obtained from the National Institute on Drug Abuse, Bethesda, MD, USA, and RTI-150 was obtained from Research Triangle Institute, Research Triangle Park, NC, USA. [11C]Cocaine and [11C]RTI-150 were synthesized using methods previously established by our laboratory.9 The chemical structures and labeling for each drug are shown in Figure 1.
CocE production
Plasmid expressing double mutant CocE (DM CocE) is described previously.6 Plasmid pET22bDMCocE was used to transform FreColi cells (RCT Technologies, Tucson, AZ, USA). Transformed cells were grown in TB media containing ampicillin antibiotic. Cells were induced with 1 mM IPTG (Sigma-Aldrich, St Louis, MO, USA) for 18 h at 18 °C and harvested at the end of 18 h. Cell pellet was lysed and CocE was purified from the lysate as described in Narasimhan et al.12 Purified protein was stored at −80 °C until use.
PET scanning
PET scanning was performed on a Siemens Focus 220 microPET scanner (Siemens, Concorde Microsystems, Knoxville, TN, USA). Animals were initially anesthetized with ketamine and Telazol, then intubated and maintained on 0.8–1.5% isoflurane for the duration of the imaging procedure. An acute venous catheter was placed in the saphenous vein for intravenous administration of the radioligand and the CocE or saline control. The catheter access was maintained with a sterile saline drip throughout the study. Animals were placed supine in the PET instrument, fitted with pulse oximetry equipment and a rectal thermistor for physiological monitoring during the procedure. Before injection of the PET radiopharmaceutical, a transmission scan was performed with a Co-57 point source to correct for attenuation in the image reconstruction. Approximately 300 MBq of the given radiopharmaceutical was injected over 5 min with an infusion pump in each animal. At 10 min post radiopharmaceutical injection, when maximal uptake was achieved, animals were challenged with either CocE (1.0 mg kg−1) dissolved in saline solution or saline alone as a control. Images were reconstructed with OSEM/MAP using measured attenuation correction, zoom factor 2, and decay corrected to the time of injection.
Data analysis
Regions of interest (ROIs) were drawn manually on each animal’s PET images fused with a rhesus MRI template over the caudate and putamen. An additional reference region was drawn over the cerebellum, which was assumed to have a negligible concentration of the dopamine transporter (DAT). The same regions were used for the saline and CocE challenges within the same animal. Regions of interest were then applied to all images to obtain time-activity curves and then normalized to the animal’s body weight and injected activity.
The pharmacokinetics of [11C]cocaine uptake into the brain were simulated using mathematical compartmental analysis commonly applied in the quantification of blood flow,13 cerebral metabolism14 and neuroreceptor binding.15 Compartments representing physiological spaces or states of [11C]cocaine in the DAT-rich regions and in the esterase challenge are assigned to the (1) blood plasma, a space representing the amount of [11C]cocaine available to cross the blood brain barrier, (2) interstitial region, a space representing the free and nonspecific bound fraction of [11C]cocaine and (3) bound state, representing [11C]cocaine that is specifically bound to the DAT. The plasma input concentration of [11C]cocaine is not explicitly known in these experiments, therefore it is estimated from the cerebellum which is assumed to have negligible specifically bound signal. The cerebellum can be modeled using only two compartments, (1) blood plasma, this is the same compartment as above and (2) reference region, representing the same compartment as interstitial region but in the cerebellum region. The use of a reference region for estimating the plasma input has been shown to be valid in the calculation of specifically bound signal in neuroreceptor studies.16 The addition of the interstitial region and bound state compartments represents the observed PET signal in the imaging studies.
Kinetic analysis of the time-activity was performed with the General Reference Tissue Model,17 a modification of the Lammertsma model allowing for the initial compartment concentrations to be nonzero. The model rate constants describe a first-order exchange between compartments using the reference region as the plasma input function. The rate constants used in the model are: R—the ratio of plasma to extracellular rate constant of the striatum and reference region (K1′/K1), k2—the extracellular to plasma rate constant in the striatum (where, k2′=k2/R in the reference region), k3—konBavail the DAT association rate (kon) times the number of available binding sites (Bavail)15 and k4—the disassociation rate from the DAT.
The model is solved by initially assigning values to the rate constants (R, k2, k3, k4), comparing the model generated time-activity curve with the PET time-activity curve, calculating the least squares sum, then iteratively adjusting the rate constants such that the least squares sum is minimized. The minimization of the least squares is done in a computer environment (IDL 6.4, ITT Visual Information Solutions, Boulder, CO, USA). An estimate of available DAT was then calculated from the solved rate constants called the binding potential, BPND=kon × Bavail/koff=k3/k4. KD (koff/kon) is assumed to be constant throughout the experiment; therefore BPND represents the number of available DAT sites for binding with [11C] cocaine.18 In addition to the compartment model, the washout rate of [11C]cocaine from the striatum was calculated using time-activity data following the peak uptake and fitting that data with an exponential function and a constant (e(−λt)+C), where λ (1 min−1) is the washout rate and C is a constant. A Mann–Whitney U test (nonparametric t-test) was used to compare statistically the washout rate between saline and CocE challenges.
Simulation data
It was hypothesized that there is a marked reduction in [11C]cocaine available in the plasma compartment to enter the brain interstitial space immediately following the esterase challenge. The pharmacokinetics of the rapid metabolism of [11C]cocaine were simulated by setting K1, the rate constant describing the influx of [11C]cocaine to the brain, to zero at 10 min post radiopharmaceutical injection and at the time of esterase injection. The influx rate constants can be estimated directly from a two-tissue compartment model if the arterial input function is known. In the present case, the input function is the cerebellum, which is assumed to have a negligible concentration of DAT and therefore represents signal from the free interstitial space and blood.
Mean time-activity curves were created for the [11C]cocaine CocE or saline control challenges by averaging the caudate and cerebellum time-activity curves from all three subjects. These averaged time-activity curves were fit with the General Reference Tissue Model to estimate the rate constants (R, k2, k3, k4). At 10 min, the DAT-rich compartment model was used to simulate the esterase challenge by assigning the compartment concentrations and rate constant calculated from the General Reference Tissue Model. In the DAT-rich compartment model, K1 was set to zero at 10 min, and was run from 10 min to 120 min to obtain the specifically bound compartment concentration over time. This simulation assumes that the introduction of the esterase compound does not alter the rate constants governing the binding of [11C]cocaine to DAT (k3) or its elimination from brain (k2 and k4).
Results
PET images for a representative subject (RGg-9) are shown in Figure 2. Note the high uptake of [11C]cocaine in the caudate and putamen, two regions of interest with high DAT density. [11C]cocaine binding diminished over 50 min following saline challenge. The reduction in [11C]cocaine binding over time was markedly accelerated following CocE challenge, indicating rapid elimination of cocaine from brain.
Sample PET images from a representative subject showing the uptake of [11C]cocaine and the subsequent elimination following saline (top) or CocE (bottom) challenge 10 min after administration of [11C]cocaine. The sequential images (from left to right) are shown in the horizontal plane. High density labeling in the caudate and putamen is easily identified. CocE, cocaine esterase; PET, positron emission tomography.
Subsequent analyses quantified the pharmacokinetics of [11C] cocaine binding. The regions of interest (caudate and putamen) and the reference region (cerebellum) were used to evaluate the time course for [11C]cocaine uptake and elimination. Time-activity curves are shown for individual subjects in Figure 3, and rate constants and BPND are reported in Table 1. The average time to peak levels of cocaine was 9.5 min, and cocaine levels dropped markedly after 40–50 min. CocE induced a significant increase in the elimination rate for cocaine (P<0.05). The elimination rate for cocaine in caudate/putamen was 3.0±0.7% per minute following saline and 8.0±1.0% per minute following CocE. Hence, CocE induced approximately a three-fold increase in cocaine elimination from brain. There was no significant difference in BPND between the saline and esterase challenges in the caudate (P=0.47) or putamen (P=0.51), indicating that CocE-induced changes in the time-activity curves and brain elimination were exclusively due to reduced cocaine concentrations in blood. In contrast, the time to peak levels of [11C]RTI-150 was considerably greater, and drug levels were sustained for the duration of the 90-min session (Figure 4 and Table 2). As predicted, CocE had no effect on the elimination rate of the cocaine analog RTI-150, which lacks the benzoyl ester linkage of cocaine.
Figure 5 shows the averaged time-activity curves and simulated model to demonstrate the consequence of K1 going to zero immediately after esterase challenge, indicating the absence of [11C]cocaine in blood for transport to the brain. Note the elimination of cocaine in the simulation was slightly faster than the measured data suggesting that [11C]cocaine was not completely eliminated from the blood at the time of CocE injection but substantially reduced compared with saline.
Averaged time–activity curves and simulated model fit for K1 set to zero. The model curve represents the simulated bound fraction of [11C]cocaine plus the cerebellum curve after esterase challenge. The decrease in the model time–activity curve is faster than the measured data suggesting that [11C]cocaine was not completely eliminated from the blood at the time of CocE injection but substantially reduced compared with the saline challenge chase. CocE, cocaine esterase.
Discussion
Cocaine overdose can result in profound cardiovascular and central nervous system alterations that are frequently lethal.19 Nevertheless, standard care for cocaine overdose primarily involves symptomatic treatment that is often insufficient.20 The usefulness of pharmacokinetic approaches to cocaine overdose has been recently suggested21 and our previous data on the pharmacokinetics of CocE in rhesus monkeys22,23 support this suggestion. This earlier work in monkeys on the kinetics of plasma cocaine elimination in the absence and presence of CocE indicated that, when administered alone, cocaine had a half-life of 50.7 min. Administration of 0.32 mg kg−1 CocE before cocaine resulted in plasma levels of cocaine that were below detection at the first measurement time of 5–7 min.22,23
The research presented here using PET neuroimaging technology demonstrated that peripheral administration of CocE removed cocaine from the rhesus-monkey brain approximately three times faster than the non-CocE control condition. CocE did not alter elimination of non-esteratic RTI-150, indicating that CocE was hydrolyzing cocaine at its ester bond, which is located between ecgonine methyl and the benzoyl moiety. Despite the rapid removal of cocaine from the circulating periphery, PET neuroimaging indicated that there were measureable levels of cocaine in the brain 20 min following CocE administration. The fact that no cocaine was detected in plasma at this time22,23 indicates that CocE’s site of action is exclusively in the periphery. Ostensibly, cocaine is removed from the brain down its concentration gradient through circulation to the periphery where it is quickly metabolized by CocE; cocaine-free blood is returned to the brain. CocE-induced changes in the rate of [11C]cocaine crossing the blood brain barrier were simulated in the current paper using PET neuroimaging and compartment modeling. The results clearly indicate that the more rapid elimination of cocaine from brain was accounted for by peripheral actions through attenuating the input function from the blood plasma compartment. This model can be adapted to other mechanisms and other enzymes, such as cocaine hydrolase24 that accomplishes similar outcomes.
The efficiency of this process is sufficient to alleviate or prevent adverse cocaine-induced central nervous system effects. Moreover, Collins et al.23 showed that CocE protected against cardiovascular effects of cocaine in rhesus monkeys. The current study indicates that protection will also be afforded to central nervous system effects at an only slightly slower rate. The results further support the development of CocE for the treatment of acute cocaine toxicity.
References
Bresler MM, Rosser SJ, Basran A, Bruce NC . Gene cloning and nucleotide sequencing and properties of a cocaine esterase from Rhodococcus sp. strain MB1. Appl Environ Microbiol 2000; 66: 904–908.
Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA et al. Crystal structure of a bacterial cocaine esterase. Nat Struct Biol 2002; 9: 17–21.
Turner JM, Larsen NA, Basran A, Barbas CF 3rd, Bruce NC et al. Biochemical characterization and structural analysis of a highly proficient cocaine esterase. Biochemistry 2002; 41: 12297–12307.
Collins GT, Brim RL, Narasimhan D, Ko MC, Sunahara RK, Zhan CG et al. Cocaine esterase prevents cocaine-induced toxicity and the ongoing intravenous self-administration of cocaine in rats. J Pharmacol Exp Ther 2009; 331: 445–455.
Cooper ZD, Narasimhan D, Sunahara RK, Mierzejewski P, Jutkiewicz EM, Larsen NA et al. Rapid and robust protection against cocaine-induced lethality in rats by the bacterial cocaine esterase. Mol Pharmacol 2006; 70: 1885–1891.
Gao D, Narasimhan DL, Macdonald J, Brim R, Ko MC, Landry DW et al. Thermostable variants of cocaine esterase for long-time protection against cocaine toxicity. Mol Pharmacol 2009; 75: 318–323.
Collins GT, Carey KA, Narasimhan D, Nichols J, Berlin AA, Lukacs NW et al. Amelioration of the cardiovascular effects of cocaine in rhesus monkeys by a long-acting mutant form of cocaine esterase. Neuropsychopharmacology 2011; 36: 1047–1059.
Jutkiewicz EM, Baladi MG, Cooper ZD, Narasimhan D, Sunahara RK, Woods JH . A bacterial cocaine esterase protects against cocaine- induced epileptogenic activity and lethality. Ann Emerg Med 2009; 54: 409–420.
Kimmel HL, Negus SS, Wilcox KM, Ewing SB, Stehouwer J, Goodman MM et al. Relationship between rate of drug uptake in brain and behavioral pharmacology of monoamine transporter inhibitors in rhesus monkeys. Pharmacol Biochem Behav 2008; 90: 453–462.
Kimmel HL, O'Connor JA, Carroll FI, Howell LL . Faster onset and dopamine transporter selectivity predict stimulant and reinforcing effects of cocaine analogs in squirrel monkeys. Pharmacol Biochem Behav 2007; 86: 45–54.
Kimmel HL, Nye JA, Voll R, Stehouwer J, Goodman MM, Votaw JR, Carroll FI, Howell LL . Simultaneous measurement of extracellular dopamine and dopamine transporter occupancy by cocaine analogs in squirrel monkeys. Synapse 2012; 66: 501–508.
Narasimhan D, Collins GT, Nance MR, Nichols J, Edwald E, Chan J et al. Subunit stabilization and polyethylene glycolation of cocaine esterase improves in vivo residence time. Mol Pharmacol 2011; 80: 1056–1065.
Kety SS . The theory and applications of the exchange of inert gas at the lungs and tissues. Pharmacol Rev 1951; 3: 1–41.
Reivich M, Kuhl D, Wolf A, Greenberg J, Phelps M, Ido T et al. The [18F]fluorodeoxyglucose method for the measurement of local cerebral glucose utilization in man. Circ Res 1979; 44: 127–137.
Mintun MA, Raichle ME, Kilbourn MR, Wooten GF, Welch MJ . A quantitative model for the in vivo assessment of drug binding sites with positron emission tomography. Ann Neurol 1984; 15: 217–227.
Lammertsma AA, Bench CJ, Hume SP, Osman S, Gunn K, Brooks DJ et al. Comparison of methods for analysis of clinical [11C]raclopride studies. J Cereb Blood Flow Metab 1996; 16: 42–52.
Votaw JR, Howell LL, Martarello L, Hoffman JM, Kilts CD, Lindsey KP et al. Measurement of dopamine transporter occupancy for multiple injections of cocaine using a single injection of [F-18]FECNT. Synapse 2002; 44: 203–210.
Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 2007; 27: 1533–1539.
Zimmerman JL . Cocaine intoxication. Crit Care Clin 2012; 28: 517–526.
Arora NP, Berk WA, Aaron CK, Williams KA . Usefulness of intravenous lipid emulsion for cardiac toxicity from cocaine overdose. Am J Cardiol 2013; 111: 445–447.
Gorelick DA . Pharmacokinetic strategies for treatment of drug overdose and addiction. Future Med Chem 2012; 4: 227–243.
Brim RL, Noon KR, Collins GT, Stein A, Nichols J, Narasimhan D et al. The fate of bacterial cocaine esterase (CocE): an in vivo study of CocE-mediated cocaine hydrolysis, CocE pharmacokinetics, and CocE elimination. J Pharmacol Exp Ther 2012; 340: 83–95.
Collins GT, Brim RL, Noon KR, Narasimhan D, Lukacs NW, Sunahara RK et al. Repeated administration of a mutant cocaine esterase: effects on plasma cocaine levels, cocaine-induced cardiovascular activity, and immune responses in rhesus monkeys. J Pharmacol Exp Ther 2012; 342: 205–213.
Zheng F, Zhan CG . Modeling of pharmacokinetics of cocaine in human reveals the feasibility for development of enzyme therapies for drugs of abuse. PLoS Comput Biol 2012; 8: e1002610.
Acknowledgements
We thank both Juliet Brown and Lisa Neidert for their expert technical assistance. These studies were supported by USPHS grants DA010344, DA031246 and the Office of Research Infrastructure Programs ODP51OD11132.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Howell, L., Nye, J., Stehouwer, J. et al. A thermostable bacterial cocaine esterase rapidly eliminates cocaine from brain in nonhuman primates. Transl Psychiatry 4, e407 (2014). https://doi.org/10.1038/tp.2014.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2014.48