Abstract
Horizontal gene transfer is recognized as an important factor in genome evolution, particularly when the newly acquired gene confers a new capability to the recipient species. We identified a gene similar to the makes caterpillars floppy (mcf1 and mcf2) insect toxin genes in Photorhabdus, bacterial symbionts of nematodes, in the genomes of the Epichloë fungi, which are intercellular symbionts of grasses. Infection by Epichloë spp. often confers insect resistance to the grass hosts, largely due to the production of fungal alkaloids. A mcf-like gene is present in all of the Epichloë genome sequences currently available but in no other fungal genomes. This suggests the Epichloë genes were derived from a single lineage-specific HGT event. Molecular dating was used to estimate the time of the HGT event at between 7.2 and 58.8 million years ago. The mcf-like coding sequence from Epichloë typhina subsp. poae was cloned and expressed in Escherichia coli. E. coli cells expressing the Mcf protein were toxic to black cutworms (Agrotis ipsilon), whereas E. coli cells containing the vector only were non-toxic. These results suggest that the Epichloëmcf-like genes may be a component, in addition to the fungal alkaloids, of the insect resistance observed in Epichloë-infected grasses.
Similar content being viewed by others
Introduction
Horizontal gene transfer (HGT) between species is well established as an important factor in genome evolution1,2. Most examples of HGT involve transfers between prokaryotic species, but the number of examples of HGT into eukaryotes is increasing3. Among the eukaryotes, the chitin cell wall of fungi is considered a structural barrier, likely reducing the frequency of HGT, but even so there are numerous instances of HGT in fungi3. Most such instances are from prokaryotes, but there are some documented fungus-to-fungus transfers. In a systematic examination of 60 fungal genomes, representing four phyla, a total of 713 genes of prokaryotic origin were detected in 53 genomes4. The Ascomycota subphylum Pezizomycotina stood out as having a large number of HGT events. Richards et al.3 analyzed the functions of 323 genes confirmed as originating from HGT into fungi and concluded that HGT played a role in expanding the nutrient acquisition and environmental colonization capacities of many fungi.
The amount of transferred DNA ranges from single genes to secondary metabolite gene clusters and even whole chromosomes5. The age of HGT events into fungi is also quite variable. For example, the transfer of a bacterial urea amidolyase gene into fungi was proposed to occur before the divergence of the subphyla Pezizomycotina and Saccharomycotina, approximately 400 million years ago (Mya)6. In contrast, transfer of the virulence gene ToxA from a fungal pathogen of wheat, Stagonospora nodorum, into the fungus Pyrenophora tritici-repentis, which resulted in the creation of a new wheat pathogen, was proposed to have occurred less than 75 years ago7.
Here we report the presence of an insect toxin gene similar to the bacterial makes caterpillars floppy genes (mcf1 and mcf2) of Photorhabdus luminescens8,9,10 in the Epichloë fungal endophytes of grasses. Epichloë are ecologically and commercially important intercellular fungal symbionts of many cool season grass species11,12. The fungi grow in between the plant cells in the above ground parts of the plant (Fig. 1). Infection by the Epichloë fungal endophytes often confers resistance to abiotic and biotic stresses13. The best understood effect of endophyte infection on the grass hosts is insect resistance, which is largely attributable to the production of toxic alkaloids by the fungi14,15. Of the hundreds of fungal genome sequences available at NCBI, only the Epichloë have a mcf-like gene, suggesting it is the result of a lineage-specific HGT event from bacteria into a predecessor of the modern Epichloë. The gene is expressed and is functional in E. typhina subsp. poae Ps1, suggesting it may be a component, in addition to the fungal alkaloids, in the observed insect resistance of some endophyte-infected grasses.
Results
Detection of a makes caterpillars floppy-like Gene in Epichloë Genomes
Recently we reported a quantitative comparative transcriptome analysis of E. festucae-infected versus endophyte-free Festuca rubra L. subsp. rubra (strong creeping red fescue) using the high throughput sequencing approach of SOLiD-SAGE16. Strong creeping red fescue is a commercially important grass species and is often naturally infected with the fungal endophyte E. festucae17,18. One SOLiD-SAGE tag recovered was from an E. festucae gene similar to the insect toxin genes makes caterpillars floppy1 and 2 (mcf1 and mcf2) of Photorhabdus luminescens8,9 and the fitD toxin genes of some Pseudomonas spp.19. Ph. luminescens is a symbiotic bacterium of entomopathogenic nematodes. When the nematodes infect their insect host, the bacteria are regurgitated into the insect hemolymph where they release several toxins that kill the insect host10,20. The mcf1 gene was discovered in a screen of Ph. luminescens cosmid clones for insect toxicity and the name comes from the effect seen after normally non-toxic Escherichia coli expressing the gene were injected into caterpillars8. mcf2, also from Ph. luminescens and fitD, from Pseudomonas protegens Pf-5 and CHA0 isolates (previously called Pseudomonas fluorescens)21 were discovered by their sequence similarity to mcf1 and have been shown to have a similar effect on caterpillars9,22. mcf1 has a 1,623 bp N-terminal extension relative to mcf2 (Fig. 2a). All of these genes encode large proteins (2,388 to 2,996 amino acids) with a TcdA/TcdB pore-forming domain similar to that of Clostridium difficile toxin B23. Mcf1 was found to kill insects by promoting apoptosis via the mitochondrial pathway20,24. The Ps. protegens FitD toxin contributes to the oral insecticidal activity of the bacteria25. The mode of action of Mcf2 is unknown9.
Gene structure of the Epichloëmcf-like genes and amino acid similarity of E. typhina subsp. poae Ps1-Mcf protein with the bacterial Mcf proteins.
(a) Diagrams of gene structure of the bacterial mcf1, mcf2, fitD and Epichloëmcf-like genes. The exons are indicated by boxes and the introns by lines. The conserved TcdA/TcdB pore-forming region is indicated by filled in boxes. The positions of the premature stop codons in some of the Epichloë genes are indicated by *. The end of intron 1 of the E. typhina subsp. poae 5819 sequence was modified from the annotated version to that of the experimentally determined position in E. typhina subsp. poae Ps1. (b) GECA analysis illustrating the amino acid sequence similarity of the E. typhina subsp. poae Ps1-Mcf protein (1,997 amino acids; accession KJ502561) to Mcf1 (2,929 amino acids; accession AF503504.2) and Mcf2 (2,388 amino acids; accession AY445665) proteins from Ph. luminescens.
The genome sequences of 13 Epichloë spp. are publicly available (15; http://www.endophyte.uky.edu/) and all contain sequences similar to the E. festucaemcf-like gene. However, similar sequences are not found in any of the hundreds of non-Epichloë fungal genome sequences currently available in NCBI. The uniqueness of the mcf-like gene to the Epichloë lineage suggests it likely arose from a HGT event from a bacterial donor. The alternative scenario in which the Epichloëmcf-like genes originated from vertical gene transmission can be rejected since this would imply that the mcf-like gene was lost in all fungal lineages except the Epichloë. Diagrams of the Epichloëmcf-like genes are shown in Fig. 2a. The annotated position of the N-terminus of the proteins was variable, but most contained genes with long open-reading frames interrupted by 2 introns. The gene structure of E. amarillans, E. baconii, E. brachyelytri, E. glyceriae, E. mollis and E. aotearoae is considered the normal structure, based on our expression analysis of the gene from E. typhina subsp. poae infecting Poa secunda subsp. juncifolia (big blue grass), designated E. typhina subsp. poae Ps126,27 (discussed below). The E. elymi, E. festucae RC (the Rose City isolate of E. festucae), E. typhina subsp. poae 5819 and E. gansuensis isolates had mutations that resulted in frame shifts and early termination codons and so would not be expected to produce an intact Mcf-like protein. The E. gansuensis var. inebrians and E. typhina E8 isolates were annotated as having N-terminal extensions relative to most of the other species. In both cases the annotated extension can be attributed to a deletion of the presumed normal coding sequence start site and recognition of upstream open reading frames as exons. The E. festucae 2368 and Fl1 isolates had deletions of the 5′ region of the gene and were annotated as beginning at a downstream region of the gene. The remarkable amino acid similarity of the predicted Mcf-like protein from E. typhina subsp. poae Ps1 to the bacterial Mcf1 and Mcf2 proteins is illustrated in Fig. 2b. The similarity extends throughout the 1,997 amino acid long protein and is particularly striking in the TcdA/TcdB pore forming domain.
The Epichloëmcf-like coding sequences were more similar in size to those of Ph. luminescensmcf2, but the overall number of amino acid identities in the region of overlap was similar between the Epichloë sequences and mcf1, mcf2 and fitD (Supplementary Table 1). The nearly identical similarity of the Epichloë sequences to all three bacterial genes, as well as a preliminary phylogenetic analysis precluded identification of the likely donor in the HGT event (discussed more below).
As is typical of bacterial genes, there are no introns in the mcf1, mcf2, or fitD genes, but there are presumed introns in the Epichloëmcf-like genes. Since it appears the Epichloëmcf-like genes originated from a HGT event, the introns must have been acquired subsequent to the HGT. Such intron acquisition has been found in other eukaryotic genes of bacterial origin4,28. Identical intron positions in genes from different recipient species suggest a single HGT event in the oldest member of the lineage, with subsequent vertical inheritance in the more recently diverged species28. All of the Epichloëmcf-like genes had an intron in the 3′ region of the gene at an identical insertion site, although there is sequence variation within the intron sequence itself. Most also had a conserved 5′ intron, except the E. gansuensis var. inebrians and E. typhina E8 isolates, both of which had deletions in the 5′ sequence, which resulted in annotated N-terminal extensions. The E. festucae 2368 and Fl1 isolates, which had truncated 5′ regions, also lacked the 5′ intron. The two identical intron positions suggest there was a single HGT insertion as well as single intron acquisition events in the original horizontally transferred mcf-like gene, which was subsequently vertically inherited throughout the speciation of the Epichloë.
Estimation of the Age of the mcf-like Gene HGT Event into Epichloë
There have been numerous phylogenetic studies of Epichloë, but none have directly addressed the order and age of species divergence. To aid in estimation of the age of the mcf-like gene HGT event into Epichloë, we undertook such an analysis. Most phylogenetic studies of Epichloë have been based on β-tubulin (tubB), elongation factor EF-1α (tefA) and γ-actin DNA sequence comparisons. In a comprehensive survey that identified optimal fungal genes for use in phylogenetic analyses, these commonly used genes were not among the best29. The two most useful genes were MCM7, a subunit of the minichromosome maintenance complex30 and TSR1, a gene required for synthesis of 40S ribosomal subunits31. MCM7 and TSR1 have been found useful in fungal phylogenies that include a wide range of taxa32,33,34. Other high-performing genes identified were NAD-dependent glutamate dehydrogenase and isoleucine tRNA synthase29.
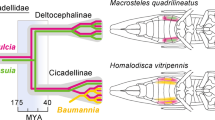
Maximum parsimony phylogenetic analyses of MCM7, TSR1, NAD-dependent glutamate dehydrogenase and isoleucine tRNA synthase sequences from 13 Epichloë, as well as some related Clavicipitaceae, are shown in Fig. 3 and Supplementary Fig. 1. Accession numbers of the sequences are given in Supplementary Table 2. To facilitate the alignments for these analyses the intron sequences were removed and only the DNA coding sequences were used. The Metarhizium anisopliae sequences were designated as the outgroups for rooting the trees since M. anisopliae is in a sister clade, which diverged prior to that of Epichloë35. In all of the trees, the two E. gansuensis sequences were in a sister clade to the rest of the Epichloë spp., which had E. glyceriae at its base. Since both E. gansuensis and E. glyceriae have the mcf-like gene in their genomes, the HGT event must therefore have occurred in the common ancestor of these two basal Epichloë spp.
Rooted 50% majority rule maximum parsimony phylogenetic tree of the MCM7 DNA coding sequences.
The Metarhizium anisopliae sequences were designated as the outgroup for rooting the tree. The numbers at the nodes are the bootstrap percentages based on 1,000 replications. The tree was based upon 2,472 total characters, of which 1,549 were constant, 343 variable characters were parsimony uninformative and 580 variable characters were parsimony informative. The unmarked nodes all had bootstrap support of 69 or higher.
To estimate the age of the HGT event we used a molecular clock approach36 similar to that used to estimate the age of polyploidy in Gossypium (cotton)37, the divergence times of maize LTR-retrotransposons38 and the age of a segmental duplication in maize39. First we estimated the divergence rate for each gene by calibration to Atkinsonella hypoxylon using the formula T = Ks/2r. A. hypoxylon (synonym: Balansia hypoxylon)40 is closely related to B. epichloë41. B. epichloë was placed in a sister clade to the clade containing E. typhina and the Claviceps spp. and the divergence of the two clades was estimated to have occurred 81 Mya35. The Ks (substitutions at synonymous sites) and Ka (substitutions at nonsynonymous sites) for the coding sequence of each gene relative to the A. hypoxylon ortholog was determined (Supplementary Table 3). In this analysis glutamate dehydrogenase from E. gansuensis var inebrians was not included, since the gene sequence was split between two contigs, which introduced a gap into the alignment. The low Ka/Ks ratios for these four genes indicate they are under purifying selection. From the Ks for each gene, the rate of divergence of synonymous sites (r) for each gene was calculated (Supplementary Table 3). The calculated rates varied from 5.93 × 10−9 to 7.72 × 10−9 substitutions per synonymous site per year, which are similar to reported fungal gene divergence rates42.
The calculated rates of divergence of each gene were then used to calculate the time of divergence (T) between the Claviceps clade and the Epichloë clade as well as between E. glyceriae and E. gansuensis sequences (Supplementary Tables 4 and 5). The mean calculated divergence time between the Claviceps clade and the Epichloë clade was 58.8 Mya and the mean divergence time between E. gansuensis and E. glyceriae was 7.2 Mya. These estimates indicate that the single introduction of the mcf-like gene into Epichloë was sometime after the divergence of the Claviceps and Epichloë clades at 58.8 Mya and before the divergence of E. gansuensis and E. glyceriae at 7.2 Mya.
The Epichloëmcf-like Gene Donor is Likely a Bacterium Related to Xenorhabdus or Photorhabdus
Phylogenetic analysis of genes derived from a HGT event can often be used to identify the donor43,44. The best matches to the Epichloë Mcf-like amino acid sequences were Mcf1 and Mcf2 from Photorhabdus luminescens and FitD from Pseudomonas spp. As discussed above, the overall similarity of the Epichloë Mcf-like sequences to the bacterial proteins was nearly the same (Supplementary Table 1). Similar Mcf2 proteins were also found in Xenorhabdus spp., also bacterial symbionts of entomopathogenic nematodes10, but have not been functionally characterized. Maximum parsimony phylogenetic analysis of the bacterial Mcf proteins and Epichloë mcf-like proteins is shown in Fig. 4. The Vibrio tubiashii sequence was chosen to root the the tree since its similarity to the Epichloë sequences is considerably lower than the Xenorhabdus, Photorhabdus and Pseudomonas sequences. Similar trees were obtained when using the the neighbor joining and branch and bound methods (Supplementary Fig. 2). The sequence alignment used to generate the tree is shown in Supplementary Fig. 3. Accession numbers of the sequences used are given in Supplementary Table 2.
Rooted 50% majority rule maximum parsimony phylogenetic tree of bacterial Mcf proteins and the Mcf-like protein sequences from E. gansuensis var. inebrians and E. typhina subsp. poae.
The Vibrio tubiashii sequence was designated as the outgroup for rooting the tree. The numbers at the nodes are the bootstrap percentages based on 1,000 replications. The tree was based upon 1,983 total characters, of which 391 were constant, 404 variable characters were parsimony uninformative and 1,188 variable characters were parsimony informative.
In the phylogenetic tree the Mcf-like sequences from E. gansuensis var. inebrians and E. typhina subsp. poae were placed as basal to the Xenorhabdus spp. Mcf2 and Photorhabdus spp. Mcf proteins, making it impossible to hypothesize as to a specific bacterial donor. Such a phylogenetic placement suggests the bacterial donor may be an unknown or extinct bacterial species related to extant Xenorhabdus or Photorhabdus spp. In a phylogenetic comparison of many Pseudomonas spp., only one clade had fitD genes and the authors suggested they were acquired through horizontal gene transfer19. The phylogenetic placement of the Pseudomonas FitD sequences as well as the overall length similarity to Mcf1 of Photorhabdus spp., are consistent with a Photorhabdus sp. as the donor to Pseudomonas.
Expression and Activity of the Epichloëmcf-like Gene
The recovery of a SOLiD-SAGE tag from Festuca rubra infected with E. festucae16 supports the expression of the E. festucaemcf-like gene. To confirm expression, we amplified and sequenced the mcf-like cDNA from Festuca rubra infected with the Rose City isolate of E. festucae, designated E. festucae RC. The expression level was extremely low and required three rounds of PCR to obtain enough sample for sequencing. Sequence analysis of the E. festucae RC mcf-like cDNA revealed a single base insertion resulting in a frame-shift and an early termination codon, relative to the E. festucae 2368 and Fl1 isolates. The single base insertion was confirmed by sequencing that region of the E. festucae RC genome. Although the mcf-like gene is expressed in the E. festucae RC isolate, it could not produce a full-length protein.
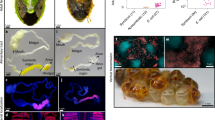
We also analyzed expression of the mcf-like gene in E. typhina subsp. poae Ps1, the fungal endophyte infecting Poa secunda subsp. juncifolia26. The E. typhina subsp. poae Ps1-mcf cDNA generated from infected plant leaf sheath tissue was easily amplified with one round of PCR (Fig. 5a). Sequence analysis of the amplified cDNA revealed it could produce a full-length protein. The cDNA sequence also confirmed the positions of the two annotated introns. The intron sequences were determined by sequencing of genomic DNA.
Expression of E. typhina subsp. poae Ps1-mcf in vivo and in E. coli.
(a) PCR product of the E. typhina subsp. poae Ps1-mcf transcript using infected Poa secunda subsp. juncifolia plant cDNA generated from oligo(dT) as template. (b) SDS-PAGE analysis of insoluble E. coli proteins. Lane 1: E. coli containing the pCold II vector only subjected to induction conditions. Lane 2: E. coli containing the pCold II::E. typhina subsp. poae Ps1-Mcf with no induction of protein expression. Lane 3: E. coli containing the pCold II::E. typhina subsp. poae Ps1-Mcf subjected to induction conditions. Arrow indicates presence of the induced protein at the expected size of 223 kD.
Since the mcf-like gene in E. typhina subsp. poae Ps1 (henceforth designated as E. typhina subsp. poae Ps1-mcf) was expressed and was predicted to produce a full-length protein, its activity against insects was assessed. The toxicity of the bacterial Mcf proteins was determined by injection of E. coli cells expressing the protein into insect hemocoel8,9,22. The E. typhina subsp. poae Ps1-mcf coding sequence was expressed in E. coli by cloning into the expression vector pCold II. This is the same strategy that was used to determine the activity of the Ps. protegens FitD protein22. A band of the expected size was seen on SDS-PAGE analysis of proteins from induced E. coli cultures (Fig. 5b). The induced protein was found in the insoluble fraction as was reported for the Ps. protegens FitD protein22. The toxicity of the induced protein was assayed by injecting black cutworms (Agrotis ipsilon) with 50 million E. coli cells harboring either the pCold II vector only or cells expressing the E. typhina subsp. poae Ps1-Mcf protein. The E. coli cells containing the vector only were not toxic to the cutworms, whereas injection of the cells expressing the Mcf protein resulted in statistically significant levels of cutworm deaths when compared to the results for the vector only controls (Table 1; Fig. 6). Lower levels of E. coli cells (20 million) expressing the E. typhina subsp. poae Ps1-Mcf protein were not toxic to the cutworms.
Toxicity of the E. typhina subsp. poae Ps1-Mcf protein to black cutworms.
Fourth instar cutworm larvae were injected with water, 50 million E. coli cells containing the pCold II vector only, or 50 million E. coli cells expressing the E. typhina subsp. poae Ps1-Mcf protein. The live cutworms curl in response to prodding.
Discussion
Here we have reported the characterization of a mcf-like gene in the Epichloë fungal endophytes of grasses that is the result of a HGT event from a bacterial donor. The gene is present in the genomes of all 13 of the Epichloë spp. for which whole genome sequence data is available, as well as 2 additional isolates used in this study, but in no other fungal species. In 11 of the 15 Epichloë sequences a long protein, containing the TcdA/TcdB pore-forming region, is predicted and in 4 of the Epichloë sequences there was a mutation leading to an early termination codon. In 3 out of these 4 cases, multiple isolates of the same species provided examples of early termination of the mcf-like gene and also of a gene capable of producing a potentially functional protein. A similar isolate specific breakdown of some alkaloid biosynthetic genes was seen among Epichloë spp.15. Phylogenetic analysis suggests the donor bacterium may have been an as yet unknown predecessor of extant Xenorhabdus or Photorabdus spp. The mechanisms underlying HGT events are not known, but clearly the donor and recipient species must be in close contact. It is easy to imagine that the recipient Epichloë sp. could have come in contact with the bacterial donor either in soil or in association with a plant host.
HGT events are of particular significance when the newly acquired gene confers a new capability to the recipient species. A recent example is the transfer of a bacterial mannanase gene to the insect Hypothenemus hampei, the coffee berry borer beetle43. The acquisition of the mannanase gene was considered to have led to the adaptation of the insect to a new host, the coffee berry, which is rich in mannans. Here we have demonstrated that injection of E. coli expressing the E. typhina subsp. poae Ps1-mcf gene into black cutworms (Agrotis ipsilon), a pest of various crops, results in death of the insect, similar to the insecticidal activities of the bacterial mcf1, mcf2, and fitD genes8,9,22. The bacterial Mcf proteins are considered critical components of the overall insect toxicity of the bacteria. The E. typhina subsp. poae Ps1-Mcf protein has been maintained as an active protein over millions of years since its acquisition by an ancestral Epichloë sp. This suggests it may also be a component, in addition to alkaloids that are well-established as having insecticidal activity15,45, in the insect resistance observed in plants containing this endophyte isolate46. Other Epichloë spp. have genes apparently capable of producing Mcf-like proteins that may also have activity against insects, as shown here for the E. typhina subsp. poae Ps1 isolate.
Bacterial toxin genes of the aerolysin family (unrelated to the mcf toxin genes) were found to have undergone recurrent HGT to many diverse eukaryotes, possibly due to their adaptive value47. Moran et al.47 proposed that genes derived from HGT are more likely to be maintained if they can function alone and are of immediate benefit to the organism. These characteristics apply to the Epichloëmcf genes, also derived from HGT, since the genes may contribute to the insect toxicity of Epichloë -infected plants.
Based on phylogenetic analyses and divergence time estimates, the single HGT event into Epichloë was estimated to have occurred sometime between 7.2 and 58.8 Mya. Dating of fungal evolution is problematic due to the minimal fossil record, so as more fungal calibration points become available it will be possible to refine the divergence time estimates presented here. The rooted phylogenetic trees generated in these analyses, based on genes established as phylogenetically high-performing genes29, are also revealing regarding the evolutionary history of Epichloë. In all four phylogenetic trees the Epichloë spp. separated into two clades, one comprised of the two E. gansuensis isolates and the other comprised of the rest of the Epichloë spp., with E. glyceriae at its base. The phylogenetic analyses presented here suggest that an as yet unknown (or possibly extinct) species was the predecessor of E. gansuensis and E. glyceriae and that the rest of the currently known Epichloë spp. were derived from E. glyceriae.
It has been proposed that Epichloë spp. evolved through coevolution with their grass hosts48. The two most basal members of this genus, E. gansuensis (host grass Achnatherum inebrians, tribe Stipeae) and E. glyceriae (host grass Glyceria striata, tribe Meliceae) are symbionts of ancient tribes of the grass subfamily Pooideae. However, E. brachyelytri, the symbiont of Brachyelytrum erectum, which is in the most ancient Pooideae tribe Brachyelytreae49,50, was phylogenetically closer to the other Epichloë spp. than to E. gansuensis or E. glyceriae. These results indicate that the speciation of Epichloë was not exclusively through coevolution with their hosts since, in an exclusive model of coevolution, E. brachyelytri would be expected to be the most basal Epichloë sp. The phylogenetic trees presented here suggest that E. brachyelytri evolved via cross species transfer of an Epichloë sp. infecting a more recently evolved host grass to the ancient plant species B. erectum. Jackson51 has also concluded that the Epichloë-host phylogenies are incongruent and do not support codivergence as a characteristic of the association. Lemaire et al.52, proposed host specificity without co-speciation for the Burkholderia bacterial leaf endosymbionts. The Epichloë – grass symbiosis may be a similar case.
Methods
Plant and Fungal Samples
Epichloë spp.-infected plants were grown by clonal propagation and maintained in 15 cm pots in the greenhouse. Each Epichloë sp. was isolated from its host plant by plating surface-sterilized leaf sheath tissue on potato dextrose agar (Difco Laboratories, Detroit, MI).
DNA and RNA Isolation
Genomic DNA of Epichloë spp. was extracted from culture grown in potato dextrose broth for 14 days on a shaker (175 rpm) at room temperature (23–25°C). The DNA was isolated as previously described53. RNA was obtained from the innermost leaf sheath tissues of Epichloë-infected plants. RNA isolation was as previously described16. Nucleic acid concentration was measured using a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Amplification and Sequencing of Epichloë mcf-like Genes and cDNAs
Initial screening to determine the presence of the mcf-like gene in E. festucae RC and E. typhina subsp. poae Ps1 fungal isolates was done using fungal genomic DNA. Upon detection of the gene, first-strand cDNA from 5 µg Epichloë-infected plant total RNA was synthesized by using SuperScript™ III Reverse Transcriptase (Life Technologies, Carlsbad, CA) and 500 ng of oligo(dT)18 primer according to the manufacturer's instructions. PCR was performed in 50 µL with either 0.5 µg of fungal genomic DNA or 1 µL Epichloë-infected plant cDNA generated from oligo(dT)18, 1.25 units of PrimeSTAR HS DNA Polymerase, 1X PrimeSTAR Buffer with Mg2+ (TaKaRa Bio Inc., Shiga, Japan), 200 µM each dNTP and 0.3 µM of each forward and reverse primer (Integrated DNA Technologies, Inc., Coralville, IA). Two-step PCR was performed by template denaturation at 98°C for 10 seconds followed by 6 minutes extension at 68°C for 30 cycles in a GeneAmp 9700 thermal cycler (Applied Biosystems, Inc., Foster City, CA). The amplification products were visualized on a 1% TBE agarose gel. The amplified genomic and cDNA PCR fragments were purified by using 0.5X Agencourt AMPure XP (Beckman Coulter, Inc., Brea, CA) to remove any fragments under 1 kb and then sequenced directly (Genewiz, Inc., South Plainfield, NJ). For each sequencing reaction, about 800 ng of purified PCR product in 10 µL was treated with 2 µL ExoSAP-IT (USB Corp., Cleveland, OH) to remove unincorporated primers and excess dNTPs. The ExoSAP-IT reaction was performed at 37°C for 15 min followed by heating at 80°C for 15 min to inactivate the enzymes.
Expression of the E. typhina subsp. poae Ps1-mcf Gene in E. coli
To test the anti-insect activity of the E. typhina subsp. poae Ps1-Mcf protein, the cDNA was cloned into the expression vector pCold II (TaKaRa Bio). The E. typhina subsp. poae Ps1 cDNA was amplified by PCR as described above with oligonucleotides that introduced a KpnI site at the 5′ end and an EcoRI site at the 3′ end. The sequences of the oligos are: forward 5′-GATATAACCATGGCTCACAACACT-3′ and reverse
5′-TCAACTGAATTCCTACTGATTTCCAGC-3′. Two hundred ng of the purified PCR product was digested with the restriction enzyme KpnI (TaKaRa Bio), purified with 0.35X Agencourt AMPure XP (Beckman Coulter, Inc) to remove any fragments under 1.5 kb, digested with EcoRI and again purified with 0.35X AMPure XP. The expression vector pCold II was similarly digested with KpnI and EcoRI. The digested pCold II plasmid was treated with shrimp alkaline phosphatase (Affymetrix, Santa Clara, CA) to prevent vector religation. Overnight ligation of the digested E. typhina subsp. poae Ps1-mcf PCR fragment and pCold II plasmid was done at a 3:1 insert:vector molar ratio using T4 DNA ligase (New England Biolabs, Inc., Ipswich, MA).
Two µL of the ligation product was used to transform 20 µl E. coli DH5α electroporation-competent cells. The transformed cells were incubated in SOC medium for 1 hour at 37°C, followed by overnight growth of cells on LB medium supplemented with 100 µg/mL ampicillin. Transformed bacterial colonies were screened for plasmids containing the E. typhina subsp. poae Ps1-mcf gene insert by using PCR as described above. Selected clones were grown in 3 mL of LB + ampicillin broth overnight followed by plasmid purification using QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). DNA sequencing of the plasmids were done as described above. A plasmid containing the correct sequence was transformed into E. coli BL21-CodonPlus (DE)-RIPL competent cells (Agilent Technologies, Santa Clara, CA). For use as a control, a transformant containing the pCold II vector only was also generated.
For expression of the E. typhina subsp. poae Ps1-Mcf protein, the BL21-CodonPlus (DE3)-RIPL transformant was induced by cold shock and isopropyl-β-D-thiogalactopyranoside (IPTG). We detected loss of E. typhina subsp. poae Ps1-mcf::pCold II recombinant plasmid in E. coli cells during the course of long induction periods. This was not observed in E. coli cells containing the pCold II vector only. Previous studies have shown that carbenicillin and ampicillin, even in high concentrations, quickly lose their ability to maintain selective pressure due to saturation of the media with the antibiotic degrading enzyme β-lactamase54,55. Extra metabolic burden placed on the bacterial cells to maintain the large recombinant plasmid combined with the steady decrease of antibiotic in culture may have contributed to the plasmid loss that we observed. In order to circumvent this problem, the culture supernatant was replaced with fresh medium at regular intervals.
To induce expression a 10 mL culture of the cells was grown for 15 h at 30°C in LB and 200 µg/mL carbenicillin. A 400 µL aliquot of the culture was pelleted, resuspended in 400 µL, added to 10 mL of fresh LB plus carbenicillin and incubated at 30°C to an OD of 0.4–0.5. The culture was then cold-shocked for 15 minutes on ice and subsequently pelleted in a Model J2-21 centrifuge (Beckman Coulter, Inc., Brea, CA) set to 5°C to obtain the bacterial cells. Fresh LB broth supplemented with carbenicillin was used to resuspend the bacterial pellets. To induce protein expression in the culture, IPTG was added to 0.1 mM and the culture was then incubated at 15°C with rotational shaking for 21–24 h. The bacterial cells were pelleted and resuspended in fresh LB and carbenicillin every 3 h for the initial 6 h and again after 12 h to prevent β-lactamase saturation in the culture supernatant. The transformant containing pCold II vector only was treated the same way.
For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, 1 ml each of E. coli cells expressing the E. typhina subsp. poae Ps1-Mcf protein and the vector only control cultures were pelleted and lysed using 1X FastBreak Cell Lysis Reagent (Promega, Madison, WI) following the manufacturer's protocol. Following lysis, samples were centrifuged at 10,000 RPM for 15 minutes and the supernatants removed. The proteins remaining in the pellet were solubilized by adding 100 µL of 2X SDS sample buffer56, boiled for 5 minutes and 30 µL was then subjected to electrophoresis on a 10% gel.
Insect Assays to Determine Toxicity of E. typhina subsp. poae Ps1-Mcf Protein
BL21-CodonPlus (DE3)-RIPL cells expressing the E. typhina subsp. poae Ps1-Mcf::pCold II recombinant insert and vector only control were prepared as described above. Cells were pelleted and resuspended in sterile water. Black cutworm (Agrotis ipsilon Hufnagel), eggs were obtained from Benzon Research Inc. (Carlisle, PA) and reared on insect diet (Southland Products Inc. Lake Village, AR) at 27°C and 14:10 (L:D) hours to fourth instars. Immediately before injection the larvae were surface sterilized in 0.8% sodium hypochlorite for 30 seconds and then rinsed in sterile distilled water. Six to ten fourth instars were injected with 5 × 107E. coli cells containing either the E. typhina subsp. poae Ps1-Mcf::pCold II induced protein or the pCold II vector only control. Additional controls were water injections and untreated samples. The assay was conducted twice. Injections were in a total volume of 5 µL using a Hamilton microsyringe with a 30-gauge needle. The larvae were then placed individually in 30 mL plastic cups with 10 g of moist (10% w/v) pasteurized (3 h at 72°C) sand with about 1 cm2 diet and incubated at room temperature (22–24°C). Mortality was assessed at 24, 48, 72 and 120 h after injection. Larvae were considered dead when they no longer reacted to being poked repeatedly with a probe. The data at the final time points were analyzed by Fisher's exact test to compare the toxin-expressing samples to the vector only controls. Significance was evaluated at P < 0.05.
Schematic Representation of mcf Gene Structure
Schematic representation of the mcf gene structures was done using Gene Structure Display Server (57; http://gsds.cbi.pku.edu.cn/index.php?input=site). Illustrative comparison of amino acid similarity for Mcf1, Mcf2 and E. typhina subsp. poae Ps1-Mcf protein was performed using Gene Evolution and Conservation Analysis (GECA) (58; http://peroxibase.toulouse.inra.fr/tools/geca_input_demo.php).
Phylogenetic Analysis
To identify genes in addition to MCM7 and TSR1 for use in the phylogenetic analyses, the FUNYBASE genome database (http://genome.jouy.inra.fr/funybase/)59 was searched for single copy orthologs identified as phylogenetically high-performing genes by Aguileta et al.29. Two genes with topological scores of 96 or above and that also had significant sequence polymorphism within Epichloë were identified, FG570 (NAD-dependent glutamate dehydrogenase) and MS444 (isoleucine tRNA synthase). DNA sequences for these genes were obtained from the Genome Project at the University of Kentucky website (http://www.endophyte.uky.edu/) or from NCBI.
The Clustal-X program60 was used to align the DNA coding sequences. The alignments generated by Clustal-X were modified manually to minimize gaps. The phylogenetic analyses were performed with the PAUP* program, version 4.0b10 for Macintosh. The phylogenetic analyses were done by using the maximum parsimony full heuristic search option set to random sequence addition, tree-bisection-reconnection (TBR) branch swapping and Multrees on, with 1,000 bootstrap replications. Gaps were treated as missing data.
The Ks and Ka values based on pairwise comparison between species were determined by using the MEGA5.2 program with the Nei-Gojobori, Jukes-Cantor model61.
Additional information
Data deposition: Sequence data for the E. typhina subsp. poae Ps1-mcf gene has been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/) under accession number KJ502561.
References
Keeling, P. J. & Palmer, J. D. Horizontal gene transfer in eukaryotic evolution. Nature Rev. Genetics 9, 605–618 (2008).
Syvanen, M. Evolutionary implications of horizontal gene transfer. Annu. Rev. Genet. 46, 341–358 (2012).
Richards, T. A., Leonard, G., Soanes, M. & Talbot, N. J. Gene transfer into the fungi. Fungal Biol. Rev. 25, 98–110 (2011).
Marcet-Houben, M. & Gabaldon, T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8 (2010).
Fitzpatrick, D. A. Horizontal gene transfer in fungi. FEMS Microbiol. Lett. 329, 1–8 (2012).
Strope, P. K., Nickerson, K. W., Harris, S. D. & Moriyama, E. N. Molecular evolution of urea amidolyase and urea carboxylase in fungi. BMC Evol. Biol. 11, 80 (2011).
Friesen, T. L. et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956 (2006).
Daborn, P. J. et al. A single Photorhabdus gene makes caterpillars floppy (mcf) allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99, 10742–10747 (2002).
Waterfield, N. R. et al. The insecticidal toxin Makes caterpillars floppy 2 (Mcf2) shows similarity to HrmA, an avirulence protein from a plant pathogen. FEMS Microbiol. Lett. 229, 265–270 (2003).
Hinchliffe, S. J., Hares, M. C., Dowling, A. J. & ffrench-Constant, R. H. Insecticidal toxins from the Photorhabdus and Xenorhabdus bacteria. Open Toxinol. J. 3, 101–118 (2010).
Clay, K. & Schardl, C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160, S99–S127 (2002).
Schardl, C. L., Scott, B., Florea, S. & Zhang, D. Epichloë endophytes: clavicipitaceous symbionts of grasses. Plant Relationships, 2nd Edition, The Mycota V [Deising, H. (ed.)] [275–306] (Springer-Verlag, Berlin Heidelberg, 2009).
Kuldau, G. & Bacon, C. Clavicipitaceous endophytes: their ability to enhance resistance of grasses to multiple stresses. Biol. Control 46, 57–71 (2008).
Schardl, C. L., Young, C. A., Faulkner, J. R., Florea, S. & Pan, J. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol. 5, 331–344 (2012).
Schardl, C. L. et al. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the Clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genetics 9, e1003323 (2013).
Ambrose, K. V. & Belanger, F. C. SOLiD-SAGE of endophyte-infected red fescue reveals numerous effects on host transcriptome and an abundance of highly expresed fungal secreted proteins. PLoS ONE 7, e53214 (2012).
Ruemmele, B. A., Wipff, J. K., Brilman, L. & Hignight, K. W. Fine-leaved Festuca species. Turfgrass Biology, Genetics and Breeding [Cassler, M. D. & Duncan, R. R. (eds.)] [129–174] (John Wiley & Sons, Inc., Hoboken, 2003).
Scott, B., Becker, Y., Becker, M. & Cartwright, G. Morphogenesis, growth and development of the grass symbiont Epichloë festucae. Morphogenesis and Pathogenicity in Fungi [Perez-Martin, J. & Di Pietro, A. (eds.)] [243–264] (Springer-Verlag, Berlin, 2012).
Loper, J. E. et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8, e1002784 (2012).
ffrench-Constant, R. H., Dowling, A. & Waterfield, N. R. Insecticidal toxins from Photorhabdus bacteria and their potential use in agriculture. Toxicon 49, 436–451 (2007).
Ramette, A. et al. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34, 180–188 (2011).
Pechy-Tarr, M. et al. Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ. Microbiol. 10, 2368–2386 (2008).
Genisyuerek, S. et al. Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol. Microbiol. 79, 1643–1654 (2011).
Dowling, A. J. et al. The insecticidal toxin Makes caterpillars floppy (Mcf) promotes apoptosis in mammalian cells. Cellular Microbiol. 6, 345–353 (2004).
Ruffner, B. et al. Oral insecticidal activity of plant-associated pseudomonads. Environ. Microbiol. 15, 751–763 (2013).
Tadych, M., Ambrose, K. V., Bergen, M. S., Belanger, F. C. & White, J. F. Jr. Taxonomic placement of Epichloë poae sp. nov. and horizontal dissemination to seedlings via conidia. Fungal Divers. 54, 117–131 (2012).
Leuchtmann, A., Bacon, C. W., Schardl, C. L., White, J. F., Jr & Tadych, M. (2014) Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 106, 202–215 (2014).
Da Lage, J.-L., Binder, M., Hua-Van, A., Janecek, S. & Casane, D. Gene make-up: rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evol. Biol. 13, 40 (2013).
Aguileta, G. et al. Assessing the performance of single-copy genes for recovering robust phylogenies. Syst. Biol. 57, 613–627 (2008).
Alabert, C. & Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell. Bio. 13, 153–167 (2012).
Gelperin, D., Horton, L., Beckman, J., Hensold, J. & Lemmon, S. K. Bms1p, a novel GTP-binding protein and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7, 1268–1283 (2001).
Schmitt, I. et al. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23, 35–40 (2009).
Raja, H. A., Schoch, C. L., Hustad, V. P., Shearer, C. A. & Miller, A. N. Testing the phylogenetic utility of MCM7 in the Ascomycota. MycoKeys 1, 63–94 (2011).
Tretter, E. D., Johnson, E. M., Wang, Y., Kandel, P. & White, M. M. Examining new phylogenetic markers to uncover the evolutionary history of early-diverging fungi: comparing MCM7, TSR1 and rRNA genes for single- and multi-gene analyses of Kickxellomycotina. Persoonia 30, 106–125 (2013).
Sung, G.-H., Poinar, G. O. Jr & Spatafora, J. W. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 49, 495–502 (2008).
Li, W.-H. Molecular Evolution [Sinauer Associates, Sunderland, 1997].
Senchina, D. S. et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20, 633–643 (2003).
Brunner, S., Fengler, K., Morgante, M., Tingey, S. & Rfalski, A. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17, 343–360 (2005).
Xu, J.-H. & Messing, J. Diverged copies of the seed regulatory Opaque-2 gene by a segmental duplication in the progenitor genome of rice, sorghum and maize. Mol. Plant 1, 760–769 (2008).
Ginns, J. Balansia hypoxylon (Ascomycota: Clavicipitaceae) in Canada: a mycosymbiont with Danthonia spicata (Poaceae). North American Fungi 6, 1–6 (2011).
Kuldau, G. A., Liu, J.-S., White, J. F., Jr, Siegel, M. R. & Schardl, C. L. Molecular systematics of Clavicipitaceae supporting monophyly of genus Epichloë and form genus Ephelis. Mycologia 89, 431–441 (1997).
Kasuga, T., White, T. J. & Taylor, J. W. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol. Biol. Evol. 19, 2318–2324 (2002).
Acuna, R. et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. USA 109, 4197–4202 (2012).
Pauchet, Y. & Heckel, D. G. The genome of the mustard leaf beetle encodes two active xylanases originally acquired from bacteria through horizontal gene transfer. Proc. R. Soc. B 280, 20131021 (2013).
Schardl, C. L. et al. Currencies of mutualisms: sources of alkaloid genes in vertically transmitted epichloae. Toxins 5, 1064–1088 (2013b).
Yue, Q., Johnson-Cicalese, J., Gianfagna, T. J. & Meyer, W. A. Alkaloid production and chinch bug resistance in endophyte-inoculated Chewings and strong creeping red fescues. J. Chem. Ecol. 26, 279–292 (2000).
Moran, Y., Fredman, D., Szczesny, P., Grynberg, M. & Technau, U. Recurrent horizontal transfer of bacterial toxin genes to eukaryotes. Mol. Biol. Evol. 29, 2223–2230 (2012).
Schardl, C. L. et al. A novel test for host-symbiont codivergence indicates ancient origin of fungal endophytes in grasses. Systematic Biol. 57, 483–498 (2008).
Kellogg, E. A. Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205 (2001).
Bouchenak-Khelladi, Y. et al. Large multi-gene phylogenetic trees of the grasses (Poaceae): progress towards complete tribal and generic level sampling. Mol. Phylogenet. Evol. 47, 488–505 (2008).
Jackson, A. P. A reconciliation analysis of host switching in plant-fungal symbioses. Evolution 58, 1909–1923 (2004).
Lemaire, B., Vandamme, P., Merckx, V., Smets, E. & Dessesin, S. Bacterial leaf symbiosis in angiosperms: host specificity without co-speciation. PLoS ONE 6, e24430 (2011).
Moy, M., Li, H., Sullivan, R., White, J. F. Jr & Belanger, F. C. Endophytic fungal β-1,6-glucanase expression in the infected host grass. Plant Physiol. 130, 1298–1308 (2002).
Korpimaki, T., Kurittu, J. & Karp, M. Surprisingly fast disappearance of β-lactam selection pressure in cultivation as detected with novel biosensing approaches. J. Microbiol. Methods 53, 37–42 (2003).
Collins, T. et al. Batch production of silk-elastin-like protein in E. coli BL21(DE3): key parameters for optimisation. Micro. Cell Fact. 12, 21 (2013).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Guo, A. Y., Zhu, Q. H., Chen, X. & Luo, J. C. GSDS: A gene structure display server. Yi Chuan 29, 1023–1026 (2007).
Fawal, N., Savelli, B., Dunand, C. & Mathé, C. GECA: a fast tool for Gene Evolution and Conservation Analysis in eukaryotic protein families. Bioinformatics 28, 1398–1399 (2012).
Marthey, S. et al. FUNYBASE: a FUNgal phYlogenomic dataBASE. BMC Bioinformatics 9, 456 (2008).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc. Acids Res. 25, 4876–4882 (1997).
Tamura, K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Acknowledgements
This work was supported with funds provided by the Rutgers Center for Turfgrass Science.
Author information
Authors and Affiliations
Contributions
K.V.A., A.M.K. and F.C.B. conceived and designed the experiments; K.V.A., A.M.K. and F.C.B. performed the experiments; A.M.K. conducted the insect injections; K.V.A., A.M.K. and F.C.B. analysed and interpreted results. All the authors discussed the results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Ambrose, K., Koppenhöfer, A. & Belanger, F. Horizontal gene transfer of a bacterial insect toxin gene into the Epichloë fungal symbionts of grasses. Sci Rep 4, 5562 (2014). https://doi.org/10.1038/srep05562
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05562
This article is cited by
-
Fungus-originated genes in the genomes of cereal and pasture grasses acquired through ancient lateral transfer
Scientific Reports (2020)
-
Differential relationship of fungal endophytic communities and metabolic profiling in the stems and roots of Ephedra sinica based on metagenomics and metabolomics
Symbiosis (2020)
-
Functional horizontal gene transfer from bacteria to eukaryotes
Nature Reviews Microbiology (2018)
-
Antibiotic and pesticide susceptibility and the Anthropocene operating space
Nature Sustainability (2018)
-
Horizontal transfer of a ß-1,6-glucanase gene from an ancestral species of fungal endophyte to a cool-season grass host
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.