Abstract
In non-small cell lung cancer (NSCLC), receptor tyrosine kinases (RTKs) stand out among causal dominant oncogenes, and the ablation of RTK signaling has emerged as a novel tailored therapeutic strategy. Nonetheless, long-term RTK inhibition leads invariably to acquired resistance, tumor recurrence and metastatic dissemination. In ALK+ cell lines, inhibition of ALK signaling was associated with coactivation of several RTKs, whose pharmacological suppression reverted the partial resistance to ALK blockade. Remarkably, ERBB2 signaling synergized with ALK and contributed to the neoplastic phenotype. Moreover, the engagement of wild-type epidermal growth factor receptor or MET receptors could sustain cell viability through early growth response 1 (EGR1) and/or Erk1/2; Akt activation and EGR1 overexpression prevented cell death induced by combined ALK/RTK inhibition. Membrane expression of ERBB2 in a subset of primary naive ALK+ NSCLC could be relevant in the clinical arena. Our data demonstrate that the neoplastic phenotype of ALK-driven NSCLC relays ‘ab initio’ on the concomitant activation of multiple RTK signals via autocrine/paracrine regulatory loops. These findings suggest that molecular and functional signatures are required in de novo lung cancer patients for the design of efficacious and multi-targeted ‘patient-specific’ therapies.
Similar content being viewed by others
Introduction
Lung tumors are the most common form of cancer (12.3%), with 1.1 million deaths worldwide, accounting for 17.8% of all cancer fatalities. Despite major diagnostic improvements and novel tailored compounds, 90% of lung cancer patients die within 1–2 years from diagnosis.1
Molecular studies have showed that nearly all lung cancers display multiple genetic defects2, 3, 4 and the abnormal activation of kinases is widely recognized as a frequent tumorigenic mechanism in non-small cell lung cancer (NSCLC).3, 5, 6, 7 Epidermal growth factor receptor (EGFR) and its ligands are commonly deregulated, leading to receptor tyrosine kinase (RTK) phosphorylation fostering cell proliferation, inhibition of apoptosis and enhancing pro-angiogenesis and invasion signals.8 Tumor-acquired EGFR mutations have been documented in 5–10% of Caucasian NSCLC, often associated to unique clinico-pathologic features.9 Specific genetic alterations of KRAS, BRAF, PIK3CA, ALK and, more recently, ROS and RET have been described in well-defined subsets of NSCLC.6, 7, 10, 11 These lesions represent ‘driver mutations’, as lung cancers depend on their constitutive activation.12, 13 Thus, molecular screening of NSCLC is mandatory to guide the most appropriate therapies (that is, small molecule kinase inhibitors).14, 15 Cancers harboring the same genetic alterations, however, respond differently to molecular targeted therapies, suggesting coexisting mutations or coactivation of other TKs.16 Finally, although molecularly targeted therapies (that is, gefitinib or erlotinib for EGFR and crizotinib for ALK fusions) are effective in NSCLC, intrinsic or acquired resistances inevitably lead to recurrence and/or metastatic dissemination. This is the case of MET activation/amplification,17, 18 or acquired somatic mutations of EGFR (T790M)19 or ALK fusions (G1269A, L1196M and ALK amplification).20, 21, 22
Chromosomal translocations involving the ALK gene have been described in 6% of NSCLC, where ALK is most frequently fused to the echinoderm microtubule-associated protein-like 4 (EML4).23 As described for other ALK chimera, the EML4–ALK fusions contain a dimerization domain, which forces the ligand-independent oligomerization of the catalytic domain of ALK, resulting in its constitutive kinase activation. ALK fusions are oncogenic ‘drivers’ imposing a full cellular transformation, and are required for the maintenance of the neoplastic phenotype.24, 25 Nevertheless, data from phase I clinical trials of ALK-rearranged NSCLC treated with the ALK inhibitor (ALKi) crizotinib showed an overall response rate of ∼60%, suggesting that synergizing or overcoming mechanisms may occur.26, 27
Here we provide novel insights into ALK-driven transformation in NSCLC, demonstrating that tumor maintenance and survival of ALK+ NSCLC cells, before any therapeutic selection, relay on the concomitant activation of multiple EGFR family RTKs. These data support the hypothesis that the degree of ALK addiction might differ ‘ab initio’ among different ALK+ NSCLC patients and its identification should provide a novel rationale for the design of efficacious ‘patient-specific’ treatments.
Results
ALK fusion proteins transform human immortalized lung epithelial cells and sustain the tumor phenotype of ALK+ NSCLC cell in vitro and in vivo
As it has been described that transformation of lung epithelial cells requires many hits, and that several oncogenes are unable to fully transform these elements, even in the presence of deregulated p53 and Rb-1 pathways, we assessed the transforming potential of ALK chimera in immortalized cell line (NuLi-1) derived from human normal airway epithelium. NuLi-1 cells were transduced with EML4–ALK variant 1 (EML4/exon13–ALK/exon20) or TFG–ALK constructs. Forced expression of EML4–ALK and TFG–ALK fusions led to constitutive phosphorylation of STAT3, SHC and Shp2 (Figure 1a), and imposed a transformed phenotype (Figure 1b).
Lung-associated ALK fusion proteins transform human immortalized lung epithelial cells and are fundamental for the maintenance of the tumor phenotype. (a) NuLi-1 cells were infected with retrovirus expressing different ALK fusion proteins and were collected 96 h after transduction. NPM–ALK and the kinase-dead (EML4–ALKK589R or TFG–ALKK231R) fusion proteins were used as controls. Total cell lysates were immunoblotted with the indicated antibodies. (b) NuLi-1 cells transduced with the indicated constructs were plated in soft agar and cultured for 3 weeks. The histograms represent the average numbers of colonies grown in soft agar from the indicated cells and constructs. The number of colonies obtained in TFG–ALK, EML4–ALK or NPM–ALK transduced cells is significantly higher as compared with untransduced cells or to cells transduced with the kinase-dead constructs (*P<0.0005; left panel). Colonies show green fluorescent protein (GFP) positivity (right panel). The number of colonies is normalized to the percentage of infection for every single construct. Cells were plated in triplicate. Results are representative of two different experiments. (c) H2228 TTA A5 and H2228 TTA A5M were cocultured with the untransduced counterpart, respectively, and grown in the presence of 1 μg/ml doxycycline. GFP expression was quantified at indicated time points by fluorescence-activated cell sorting (FACS) analysis and normalized against the initial time point (day 0). Results are representative of two different experiments. (d) H3122, H2228 and H1395 cell lines were treated every 24 h with 300 nM CEP-14083 or CEP-26939. Tetrametylrodamine methyl ester staining followed by FACS analysis was performed after 48 and 72 h of treatment to check the percentage of apoptotic cells. Significance according to the analysis of variance test corresponded to *P<0.05 and **P<0.005. Results are means of duplicates (s.e.m.), and graphics show one representative experiment from two independent assays.
To evaluate the biological effects of EML4–ALK, we transduced ALK-positive cell lines (H3122 and H2228) with ALK-specific inducible short hairpin RNA (shA5) or control short hairpin RNA (shA5M) lentivirus cassettes. Alternatively, cells (ALK+ and ALK− (H1395)) were treated with ALK-selective inhibitors (CEP-14083 and CEP-26939, from Cephalon/Teva, West Chester, PA, USA).28, 29 ALK inhibition led to a significant decrease in the phosphorylation of known ALK downstream targets (STAT3, Erk1/2, Shp2 and Akt; Supplementary Figure 1A).24 As expected, ablation of ALK signaling was associated with growth impairment (Figure 1c and Supplementary Figure 1B) and a very high rate of apoptosis for the H3122 cells (Figure 1d and Supplementary Figure 1C). Consistently, in vivo treatment with CEP-26939 led to a decreased number of proliferating cells, without any significant loss of ALK protein expression (Supplementary Figures 1D and E), and to tumor mass regression. This treatment, however, did not result in tumor eradication (Supplementary Figure 1F).
Signaling through the EGFR–TK proteins contributes to the survival and growth of ALK+ NSCLC cells
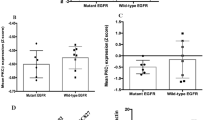
On the basis of our previous phosphoproteomics findings30 and on the different degree of ALK oncogenic addiction of ALK+ NSCLC lines and primary tumors,31 we investigated the association of anti-ALKi29 with small anti-TK drugs currently into the clinics. Combined treatment of H2228 cells with ALKi and with the EGFR inhibitor gefitinib, or with an anti-EGFR monoclonal antibody (cetuximab), resulted in increased cell death. Similar findings were seen in H3122 cells after single anti-ALK treatment (Figure 2a and Supplementary Figures 2A and B), possibly related to higher protein expression and phosphorylation status of EGFR in only H2228 cells (Supplementary Figures 2C and D). Importantly, H2228 and H3122 cells were both insensitive to single treatment with gefitinib. As other members of the EGFR family are associated to RTK inhibitor resistance, we then investigated the functional role of ERBB2 and ERBB3 in the less-responsive H2228 cells.18, 20 We observed a marked presence of ERBB2/ERBB3 and EGFR/ERBB3 heterodimers in H2228 in the absence of exogenous stimuli (Supplementary Figure 3A). Moreover, ERBB3 protein/mRNA levels were upregulated in H2228 when treated with anti-ALK small molecules (Figure 2b). Interestingly, we detected a constitutive phosphorylation of ERBB2 that was completely abrogated upon treatment with lapatinib (Figure 2b and Supplementary Figure 3B), and the combination of lapatinib with CEP-28122 suppressed cell growth and significantly reduced the number of viable cells as compared with CEP-28122 alone (Figure 2c). In contrast, the combined treatment had no further effect in H3122 cells (Supplementary Figure 3C). Remarkably, a significant subset of primary ALK+ NSCLC (12/21, 57%; but ALK− NSCLC 38/180, 21%) shows a strong immunohistochemical membrane staining for ERBB2 (Figure 2d).
In H2228, apoptosis is increased by the combined treatment with ALK inhibitors and gefitinib or lapatinib. (a) H2228 cells were treated in 0.05% FCS with CEP-14083 or CEP-28122 (300 nM), and gefinitib (G; 1 μM) as single agent or in combination. Apoptosis was measured by tetrametylrodamine methyl ester (TMRM) staining and fluorescence-activated cell sorting (FACS) analysis at the indicated tome points. *Significance is referred to the corresponding single-agent treatment. (b) H2228 cells were treated with CEP-28122 alone or in combination with lapatinib (L) or gefitinib (G) in 0.05% FCS for 24 h. Total cell lysates were blotted with the indicated antibodies. (c) H2228 cells were treated with CEP-28122 alone or in combination with lapatinib in 0.05% FCS. TMRM staining followed by FACS analysis was performed at indicated times of treatment to check the percentage of apoptotic cells. (d) Primary ALK+ NSCLC were stained with anti-ERBB2 or anti-ALK antibodies. Shown are two representative cases.
Overall, these data suggest that ALK inhibition might be overcome by alternative RTK pathways in ALK+ NSCLC cells.
Ligand-mediated activation of tyrosine receptor kinases rescues ALK kinase inhibition through AKT and Erk1/2 pathways
As EGF can be secreted by lung epithelial cancer and/or stromal cells, we investigated whether an autocrine–paracrine EGF-mediated activation of the EGFR signaling in H2228 cells could explain their partial resistance to ALK inhibition and overcome ALK inhibition of ALK-sensitive cells. Consistent with recently published data, upon stimulation with exogenous EGF the effects associated with the abrogation of the ALK signaling (CEP-28122) were overcome in both cell lines as documented by an increased cell viability and growth, which were restored to baseline levels (Figure 3a and Supplementary Figures 4A and B). Cells cotreated with gefitinib and/or expressing a mutant form of the EGFR (T790, data not show) could not be rescued by the exogenous EGF (Supplementary Figure 4A).20, 32
EGF or Heregulin stimulation rescues ALK-treated NSCLC cells through Erk1/2 and AKT. (a) Cell viability of treated H2228 and H3122 cells with or without EGF stimulation (10 ng/ml) was evaluated with Cell Titer-Glo Luminescent cell viability assay at the indicated time points. Significances * and ** is referred to the corresponding single-agent treatment. (b) H2228 and H3122 cells were treated for 6 h with 300 nM CEP-28122, alone or in combination with 1 μM gefitinib (G) in 0.05% FCS. Cells were stimulated with EGF (10 ng/ml) for 15 min and then collected. Total cell lysates were blotted with the indicated antibodies. (c,d) H2228 cells were treated with CEP-28122 alone or combined with gefitinib or lapatinib for 24 h, and with or without Heregulin (HRG) stimulation (50 ng/μl) for 30 min and then collected. Total cell lysates were blotted with the indicated antibodies (c). Cell viability was evaluated as previously described at the indicated time points. Significance according to Student’s t-test corresponded to *P<0.05 (d). Significances * and ** are referred to the corresponding single treatment without EGF or Heregulin stimulation. Results are representative of three different experiments.
The molecular mechanisms operating in the EGF/EGFR-mediated rescue were first examined by the phosphorylation status of ALK and known ALK effectors. These were not affected. On the contrary, EGF stimulation could sustain the phosphorylated status of EGFR, Akt and Erk1/2, despite an ALK inhibition (Figure 3b). Finally, an increased expression of the proapoptotic protein BIM was seen only with the dual inhibitor cotreatment in H2228, whereas ALK inhibition alone was sufficient to induce a significant upregulation of BIM in H3122 (Figure 3b).33 Similar results were obtained by heregulin stimulation. Upon heregulin stimulation the phosphorylation of ERBB3, Erk1/2 and Akt significantly increased (Figure 3c and Supplementary Figure 4B) and cell viability was rescued despite ALK inhibition (Figure 3d).
As hepatocyte growth factor (HGF)/Met signaling or Met overamplification can be involved in gefitinib resistance in lung cancer,18, 34 we then explored the effects of HGF. HGF promoted a moderate proliferation and sustained cell viability upon ALK inhibition through Erk1/2 and Akt pathways (Supplementary Figures 5A and B). As expected, crizotinib, a dual ALK–Met inhibitor, abolished the prosurvival signals triggered by HGF (Supplementary Figure 5C).
As Src has a pathogenetic role in EGFR-mutated NSLCL35 and is also a critical interactor in ALK+ anaplastic large cell lymphoma,24 we applied a selective Src kinase inhibitor-1 as a single agent (Figure 4a) or in combination with CEP-28122. The combined treatment increased cell death only in H2228 that was partially rescued by EGF administration (Figures 4b–e). These data suggest that Src-independent pathways might be active in H2228 cells. On the contrary, EGF did not restore cell viability of H3122 cells treated with ALK and Src–TK inhibitor. Similar findings were observed using dasatinib, a pan Src inhibitor (data not shown).
Src is a common node downstream of ALK and EGFR signaling pathways in NSCLC ALK-positive cells. (a) H2228 and H3122 cells were treated with CEP-28122 and with 5 μM SKI-I (S) in 0.05% FCS for 24 h and for 12 h, respectively. Total cell lysates were blotted with the indicated antibodies. (b–e) H2228 and H3122 cells were treated with CEP-28122 alone or in combination with 5 μM Src kinase inhibitor-1 (SKI-1), and with or without EGF stimulation (10 ng/ml) every 24 h. The cell proliferation rate was evaluated with the Cell Titer-Glo Luminescent cell viability assay (b, d) and the percentage of apoptotic cells was quantified by tetrametylrodamine methyl ester staining followed by fluorescence-activated cell sorting analysis (c, e).
Early growth response 1 (EGR1) gene is upregulated by active ALK and EGFR, and promotes survival
To identify ALK cooperating oncogene(s) in H2228 NSCLC cells, we interrogated their transcriptome before and after TK inhibitor treatment. Gene expression analysis showed that a single gene, early growth response 1 (EGR1), was significantly suppressed by the combined ALK/EGFR treatment (fold-change >2, P<0.001; Supplementary Figure 6). Quantitative reverse transcription–PCR confirmed that the loss of ALK signaling or the dual treatment led to a substantial inhibition of the EGR1 expression (Figure 5a), whereas gefitinib had no effect. Downregulation of EGR1 could be inhibited by exogenous EGF that overcame cell death as well (Supplementary Figure 7A). Moreover, Src inhibition in association with CEP-28122 strongly reduced the level of expression of EGR1 in both cell lines (Supplementary Figure 7B). Reversely, ectopic expression of EGR1 in H2228 cells (Supplementary Figure 7C) prevented cell growth inhibition and cell death via CEP-28122 and/or gefitinib combination (Figures 5b and c). Finally, forced expression of EGR1 could revert BIM levels to baseline (Figure 5d and Supplementary Figure 7D), meanwhile knockdown of EGR1 in H2228 through short hairpin RNA impaired cell growth (data not shown). Overall, these findings indicate that both ALK and EGFR signaling are required for EGR1 expression and demonstrated a central role of this transcription factor in ALK+ NSCLC cells.
EGR1 is regulated by ALK and EGFR, and promotes survival of H2228 cells. (a) H2228 and H3122 cells were treated with CEP-28122 and/or gefitinib (G) in 0.05% FCS for 24–48 h and 6–12 h, respectively. Total RNA was extracted and analyzed for EGR1 expression by real-time PCR. Significance according to the t-test corresponded to *P<0.05 and **P<0.005. (b, c) H2228 (mock) and H2228 transduced with EGR1 cDNA (pWPI-EGR1) were treated with CEP-28122 and G, in single or in combination in 0.05% FCS for 96 h and 120 h, respectively. Cell viability was evaluated by Cell Titer-Glo Luminescent kit (b) and the percentage of apoptotic cells was measured by tetrametylrodamine methyl ester staining followed by fluorescence-activated cell sorting analysis (c). Significances * and ** are referred to the corresponding untransduced cells (mock). Results are representative of three different experiments. (d) Cells treated with CEP-28122 and/or G in 0.05% FCS for 24 h were collected for western blot analysis with the indicated antibodies.
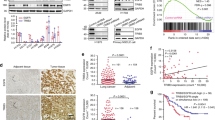
To identify EGR1-regulated genes, we investigated the transcriptional profile of H2228 cells stably transduced with EGR1 (EGR1+). This revealed a set of 780 genes linked to EGR1 (fold-change >2, P<0.001). Notably, EGR1 overexpression resulted in ERBB3 mRNA upregulation that was further increased by ALK inhibition (Supplementary Figure 8). Expression profiling of EGR1+ cells treated with ALKi and EGFR–TK inhibitor reported two clusters of genes behaving independently of ALK inhibition, as these genes showed a marked up- or downregulation in wild-type treated cells, and no change or even an opposite behavior in EGR1+ cells (Figure 6). In particular, seven genes were upregulated by EGR1 transduction and remained significantly upregulated despite the treatment with TK inhibitor, suggesting direct and robust regulation by this transcription factor (that is, TFF2, PPDPF, AGR2 and CCL5; Supplementary Figure 8).
EGR1 overexpression regulates the transcription of genes independently of ALK inhibition. (a) Heat map of gene expression signature in response to ALK and/or EGFR inhibition in H2228 cells and in H2228 cells overexpressing EGR1. The color scale bar represents relative gene expression changes calculated as log2 (inhibited/non-inhibited). (b) List and expression changes of genes up- or downregualated by the overexpression of EGR1 independently of ALK or EGFR inhibition.
Discussion
The discovery of specific genetic lesions in lung cancer patients has profoundly changed their clinical management. Kinases are now recognized as critical tumorigenic events in human cancer. Constitutively active receptor and non-RTKs are excellent examples for the development of modern ‘patient-specific’ and/or ‘tumor-restricted’ therapies.36 However, clinical responses are heterogeneous as a result of multiple mechanisms of resistance. It is believed that multi-targeted drugs may be more appropriate than highly restricted inhibitors. In NSCLC patients, ALK is a powerful driver/oncogene;37 nevertheless, crizotinib, a dual ALK/Met inhibitor, is successful only in a subset of ALK+ patients (∼60%). Moreover, responder patients inevitably become refractory.38 Thus, the different responses to ALKi may underlie intrinsic mechanisms that undermine ‘ab initio’ their efficacy and/or favor the occurrence of resistance. Their identification provide the basis for a molecular stratification of ALK+ NSCLC patients and drive the selection of more compliant treatments.21, 39
Here we studied the level of ALK oncogenic addiction of human ALK+ NSCLC and the cooperative role of several kinases. We show that the different responses of NSCLC cell lines to ALK are dependent on the activation of alternative pathways in which wild-type RTK can overcome ALK signaling blockade.
Using phosphoproteomics, different studies have demonstrated that lung cancer cells and primary tumors express kinases able to jointly participate to hierarchical networks or independently signal through separate parallel pathways.5, 40 Our previous phosphomapping findings showed that ALK inhibition is rapidly followed by the parallel activation/phosphorylation of several other TKs (EGFR, Met, fibroblast growth factor receptor, Jak1 or insulin-like growth factor receptor) in H2228 cells, but not in H3122 cells, which are markedly ALK-addicted.30 Interestingly, the dual inhibition with gefitinib or cetuximab overcomes this resistance feedback loop and increases cell death. The elevated levels of EGFR protein in H2228 compared with H3122, without displaying any amplification or known mutation, might explain their partial addiction to ALK and the better responsiveness to the cotreatment. Notably, other members of the EGFR family, ERBB2 and ERBB3, can also overcome the partial resistance to ALKi, as the combination with lapatinib and an ALKi markedly reduced ERBB2 phosphorylation and cell viability. This suggests that forced EGFR family signaling (EGFR and ERBB2), even in naive/untreated NSCLC cells, may be critical, similar to what has been recently described for the acquired resistance to ALKi.20, 41 Nevertheless, we demonstrated that this occurs in a context of wild-type EGFR and non-mutated ALK and, more importantly, in absence of drug selection.17, 42 Our studies are in line with those described by Prahallad et al.43, which have documented an intrinsic resistance to vemurafenib caused by a feedback activation of EGFR signaling in BRAF (V600E) mutant colon cancer cells. Thus, EGFR members can elicit an undermining mechanism that sustains the partial resistance seen in some ALK+ NSCLC patients, or ultimately post-therapy resistance to ALKi.41 A greater complexity is provided by the observation that the engagement of wild-type EGFR or wild-type MET receptors could also restore cell viability of H2228 and H3122 cells via EGR1 and/or Erk1/2 and Akt activation.32 Remarkably, in H2228 the effects of HGF on cell survival were less significant, more likely for the partial dependence of these cells on EGFR signaling. We believe that EGFR member family expression levels or phosphorylation should be determined at diagnosis in order to benefit the most on combined therapies.
As cancer cells often trigger alternative pathways to bypass specific RTK inhibition, we further dissected other signaling pathways to find a common node of ALK and other RTKs. In ALK+ NSCLC cells, Src kinase represents a suitable target as in ALK+ anaplastic lymphoma its ablation has relevant effects in analogy to many human cancers.35 In H2228 cells, Src inhibitors alone or in combination with ALKi increased the percentage of apoptotic cells similarly to ALK and gefitinib, suggesting that Src mediates ALK and EGFR signaling in NSCLC. These data are consistent with the notion that ALK and EGFR pathways functionally compensate each other. Nevertheless, other alternative signaling pathways might be operational in H2228, as EGF partially recovered cells from death. Indeed, rescue by EGF in H3122 was completely abrogated upon ALK and Src inhibition, suggesting that EGF/EGFR signaling triggers prosurvival responses through Src.
Next, to define additional putative downstream ‘culprits’, we used gene expression profiling analyses, which showed the EGR1 gene as a key regulator. The cotreatment with ALKi and gefitinib downregulated the expression of a single transcript, EGR1. EGR1 is a transcription factor, known to have a polyhedric/versatile role in human cancers,44 engaged in proapoptotic or prosurvival signals, depending on the cellular context.45 In solid tumors, including lung cancers, EGR1 is a downstream target of EGFR through the mitogen-activated protein kinase signaling pathways.46 Indeed, its overexpression, able to sustain cell viability in presence of ALK and EGFR inhibitors, was characterized by well-defined transcriptional signature. Among the upregulated genes, TFF2 (trefoil factor 2) and AGR2 (anterior gradient 2) seem to be relevant, as in human cancers they contribute to tumor growth, resistance to apoptosis and cell migration.47, 48 Thus, EGR1 could represent an ideal target for the design and implementation of new therapeutic approaches in ALK-positive NSCLC.
Our data indicate that the molecular characterization of primary lung cancers is required to foresee whether any given therapeutic protocol may be successful. The genetic heterogeneity of lung tumors implies that parallel pathways or different mechanisms of resistance may coexist ‘ab initio’ and that targeting a single molecule might be ineffective. This stratification is necessary to develop new combined treatment for NSCLC patients. Targeting common molecules downstream of RTKs, such as EGR1 or Src, could bypass intrinsic or acquired drug resistance derived from the treatment with a single RTK inhibitor. Our data support the idea that the development of less-restricted drugs or the design of new therapeutic protocols making use of multiple TK inhibition might be more efficacious, even for the first-line treatment of NSCLC.
Materials and methods
Cell cultures, inhibitors and growth factors
Human lung cancer EML4–ALK-positive cell lines, H2228 (var 2: EML4/es6–ALK/es20) and H3122 (var 1: EML4/es13–ALK/es20), were grown in RPMI medium (Lonza, Basel, Switzerland) with 10% fetal calf serum (FCS). Human HEK-293T cells were cultured in Dulbecco’s modified Eagle’s medium with 10% FCS. Human immortalized airway epithelial cells (NuLi-1) were grown in bronchial epithelial cell basal medium (Lonza) with 10% FCS on human collagen type-VI-coated plates (Sigma, St Louis, MO, USA).
ALK inhibitors, CEP-14083, CEP-26939 and CEP-28122 (Cephalon), were used at 300 nM/l; gefitinib (ZD1839, Iressa) at 1 μM/l; lapatinib at 2 μM/l; Src kinase inhibitor-1 (Calbiochem, Millipore Corporation, Billerica, MA, USA) at 5 μM/l; and PHA-665752 (Met inhibitor) at 250 nM. EGF was used at 10 ng/ml, Heregulin was used at 20 ng/ml, and HGF (Sigma) was used at 50 ng/ml.
Cell proliferation, cell cycle analysis, cell apoptosis and clonogenic assays
In 24-well plates, 8 × 104 cells/ml were grown in duplicates. The treatment with kinase inhibitors or with growth factors was done every 24 h in 0.05% FCS. Cell growth was analyzed using Cell Titer-GloMax assay (Promega, Fitchburg, WI, USA). For cell cycle analysis, cells were fixed for 1 h in 70% ethanol at 4 °C. After washing, cells were treated with RNase (0.25 mg/ml) and stained with propidium iodide (50 μg/ml). The sub-G0-phase fraction was calculated using the Modfit program from Becton-Dickinson (San Jose, CA, USA). Apoptosis was measured by flow cytometry after staining with the mitochondrion-permeable voltage sensitive dye tetrametylrodamine methyl ester and fluorescence-activated cell sorting analysis. Clonogenic assay, were performed as described.28
DNA constructs
EGR1 cDNA in pCMV-Sport6 was purchased from ImaGenes GmbH (Berlin, Germany). EGR1 cDNA was then subcloned into the lentiviral vector pWPI for lentivirus production. pCMV-Sport6 containing EGR1 construct was digested to obtain a EcoRV/NotI fragment, blunted with Klenow enzyme (Roche, Basilea, Switzerland) and cloned into the PmeI site of the lentiviral vector pWPI.
Lentivirus and short hairpin RNA production, cell infection and cell sorting
Lentivirus particles were obtained as described.27 For cell sorting enrichments, EGFP-positive cells were sorted using a MoFlo High-Performance instrument (DAKO Cytomation, Glostrup, Denmark) and EGFP expression was checked by flow cytometry over time.
Cell lysis and western blot analysis
H3122 and H2228 were plated on six-well plates and treated with specific kinase inhibitors in Dulbecco’s modified Eagle’s medium with 0.05% FCS. Cells were lysed with GST-FISH (10 mM MgCl2, 150 mM NaCl, 1% NP40, 2% Glycerol, 1 mM EDTA, 25 mM HEPES (pH 7.5)) buffer supplemented with protease inhibitors (NaF, phenylmethanesulfonylfluoride, PIC and Na3VO4) by scraping. The list of the antibodies is provided in Supplementary Information.
RNA extraction, cDNA synthesis and reverse transcription–PCR
Total RNA was isolated from cells with TRIZOL lysis solution (Invitrogen, Life Technologies Ltd, Paisley, UK). One microgram of total RNA was treated with 5 U/μl of DNase I recombinant RNase free (Roche) and it was subjected to reverse transcription with SuperScript III reverse transcriptase (Invitrogen). The cDNA obtained was subjected to PCR using specific primers for PBGD, B2M and ABL as control of cDNA quality.
cDNA microarray and analysis
Total RNA was extracted using the TRIZOL reagent (Invitrogen) and purified using the RNeasy total RNA Isolation Kit (Qiagen, Hilden, Germany). RNA integrity was verified by means of a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Hybridization was performed on HumanWG-12v4 BeadChips (Illumina, San Diego, CA, USA), using biological triplicates for each condition. cDNA and biotinylated cRNA were generated by Illumina TotalPrep RNA Amplification Kit (Ambion, Life Technologies Ltd, Paisley, UK). Data were processed with the Illumina GenomeStudio software using the following thresholds for significant differential expression: ‘Illumina Custom’ test P-value <0.001, fold-change >2. Gene expression data were clustered and visualized with the GEDAS software.29
Real-time quantitative PCR
cDNA was transcribed using SuperSCRIPT III following the manufacturer’s instructions (Invitrogen). Quantitative reverse transcription–PCR was performed in triplicate on ICycler iQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with SYBR green dye.
Immunofluorescence and immunohistochemistry
Cells were grown for 12 h on glass coverlips, fixed in phosphate-buffered saline containing 4% paraformaldehyde at room temperature (10 min), and then permeabilized with phosphate-buffered saline containing 0.3% Triton X-100 for 5 min. Anti-EGFR (1:100, Cell Signaling Technology, Danvers, MA, USA), ERBB2 (1:300, Dako) and ERBB3 (1:100, LabVision, ThermoScientific, Kalamazoo, MI, USA) were incubated O/N, and heterodimer formation was detected using DUOLINK II assay (Olink Bioscience, Uppsala, Sweden). Images were acquired by Imager.Z1 fluorescent microscope (Zeiss) and analysed with the Isis and Metaphor software. Immunohistochemistry was performed on a semi-automated immunostainer as described previously.30 The following antibodies were used: anti-ALK (clone D5F3; Cell Signaling Technology) and anti-ERBB2 (clone CB11; Novocastra, Leica, Milano, Italy).
Mice and in vivo bioluminescence experiments
CB/17 scid and NOD-SCID mice (Charles River Laboratories Italia S.p.A., Wilmington, DE, USA) were inoculated subcutaneously in both flanks (single-cell suspension 1 × 107 in 0.2 ml phosphate-buffered saline). Mice were treated twice a day with 100 mg/kg of Comp-2. Tumor growth was measured over time. For imaging studies, luciferase-positive cell xenografts were analyzed in vivo by IVIS100 (Xenogen, Grantham, Lincolnshire, UK) instrument after intraperitoneal injection with D-luciferin (10 μl/g body weight, 15 mg/ml). Mice were handled and treated in accordance with the European Community guidelines.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006; 439: 353–357.
Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446: 153–158.
Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007; 450: 893–898.
Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007; 131: 1190–1203.
Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012; 18: 375–377.
Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012; 18: 382–384.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Jackman DM, Chirieac LR, Janne PA . Bronchioloalveolar carcinoma: a review of the epidemiology, pathology, and treatment. Semin Respir Crit Care Med 2005; 26: 342–352.
Sekido Y, Fong KM, Minna JD . Molecular genetics of lung cancer. Annu Rev Med 2003; 54: 73–87.
Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012; 18: 378–381.
Pao W, Girard N . New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011; 12: 175–180.
Haber DA, Settleman J . Cancer: drivers and passengers. Nature 2007; 446: 145–146.
Pao W, Kris MG, Iafrate AJ, Ladanyi M, Janne PA, Wistuba II et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res 2009; 15: 5317–5322.
Sequist LV, Heist RS, Shaw AT, Fidias P, Rosovsky R, Temel JS et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011; 22: 2616–2624.
Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 2007; 318: 287–290.
Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010; 17: 77–88.
Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007; 316: 1039–1043.
Engelman JA, Janne PA . Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008; 14: 2895–2899.
Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012; 4: 120ra117.
Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012; 18: 1472–1482.
Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010; 363: 1734–1739.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566.
Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G . The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer 2008; 8: 11–23.
Piva R, Chiarle R, Manazza AD, Taulli R, Simmons W, Ambrogio C et al. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood 2006; 107: 689–697.
Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol 2011; 12: 1004–1012.
Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010; 363: 1693–1703.
Ott GR, Wells GJ, Thieu TV, Quail MR, Lisko JG, Mesaros EF et al. 2,7-disubstituted-pyrrolo[2,1-f][1,2,4]triazines: new variant of an old template and application to the discovery of anaplastic lymphoma kinase (ALK) inhibitors with in vivo antitumor activity. J Med Chem 2011; 54: 6328–6341.
Cheng M, Quail MR, Gingrich DE, Ott GR, Lu L, Wan W et al. CEP-28122, a highly potent and selective orally active inhibitor of anaplastic lymphoma kinase with antitumor activity in experimental models of human cancers. Mol Cancer Ther 2012; 11: 670–679.
Voena C, Panizza E, D’Amico L, Ambrogio C, Martinengo C, Boccalatte FE et al. EML4-ALK signaling is required for the maintenance of neoplastic phenotype of non-small cell lung cancer cells: novel strategy for lung cancer tailored therapies. AACR 101ST Annual Meeting 2010. American Association of Cancer Research (AACR), Washington DC, USA, 2010.
Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009; 27: 4247–4253.
Yamada T, Takeuchi S, Nakade J, Kita K, Nakagawa T, Nanjo S et al. Paracrine receptor activation by microenvironment triggers bypass survival signals and ALK inhibitor-resistance in EML4-ALK lung cancer cells. Clin Cancer Res 2012; 18: 3592–3602.
Takezawa K, Okamoto I, Nishio K, Janne PA, Nakagawa K . Role of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancer. Clin Cancer Res 2011; 17: 2140–2148.
Yano S, Wang W, Li Q, Matsumoto K, Sakurama H, Nakamura T et al. Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor-activating mutations. Cancer Res 2008; 68: 9479–9487.
Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med 2011; 17: 461–469.
Janku F, Stewart DJ, Kurzrock R . Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat Rev Clin Oncol 2011; 7: 401–414.
Gerber DE, Minna JD . ALK inhibition for non-small cell lung cancer: from discovery to therapy in record time. Cancer Cell 2010; 18: 548–551.
Sasaki T, Janne PA . New strategies for treatment of ALK-rearranged non-small cell lung cancers. Clin Cancer Res 2011; 17: 7213–7218.
Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci USA 2011; 108: 7535–7540.
Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA 2008; 105: 692–697.
Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res 2011; 71: 6051–6060.
McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J . Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res 2010; 70: 1625–1634.
Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012; 483: 100–103.
Sauer L, Gitenay D, Vo C, Baron VT . Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene 2010; 29: 2628–2637.
Thiel G, Cibelli G . Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol 2002; 193: 287–292.
Uramoto H, Shimokawa H, Hanagiri T, Kuwano M, Ono M . Expression of selected gene for acquired drug resistance to EGFR-TKI in lung adenocarcinoma. Lung Cancer 2011; 73: 361–365.
Park K, Chung YJ, So H, Kim K, Park J, Oh M et al. AGR2, a mucinous ovarian cancer marker, promotes cell proliferation and migration. Exp Mol Med 2011; 43: 91–100.
Kosriwong K, Menheniott TR, Giraud AS, Jearanaikoon P, Sripa B, Limpaiboon T . Trefoil factors: tumor progression markers and mitogens via EGFR/MAPK activation in cholangiocarcinoma. World J Gastroenterol 2011; 17: 1631–1641.
Acknowledgements
We thank Dr Paola Bernabei for the Flow Cytometry studies and Cell Sorting, Dr Flavio Cristofani for the mouse housing and care, and Dr Daniela Cantarella for microarray-based gene expression profiling. We thank Dr Klingelhutz J for the immortalized airway epithelial cells (NuLi-1). This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC No. IG 9196, Milan, Italy), Ministero dell’Università e Ricerca Scientifica and Regione Piemonte and Compagnia di San Paolo, Torino (Progetto Oncologia).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Voena, C., Di Giacomo, F., Panizza, E. et al. The EGFR family members sustain the neoplastic phenotype of ALK+ lung adenocarcinoma via EGR1. Oncogenesis 2, e43 (2013). https://doi.org/10.1038/oncsis.2013.7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2013.7