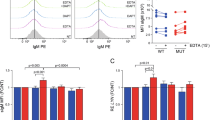
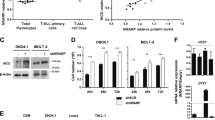
Abstract
Notch receptors have been implicated as oncogenic drivers in several cancers, the most notable example being NOTCH1 in T-cell acute lymphoblastic leukemia (T-ALL). To characterize the role of activated NOTCH3 in cancer, we generated an antibody that detects the neo-epitope created upon gamma-secretase cleavage of NOTCH3 to release its intracellular domain (ICD3), and sequenced the negative regulatory region (NRR) and PEST (proline, glutamate, serine, threonine) domain coding regions of NOTCH3 in a panel of cell lines. We also characterize NOTCH3 tumor-associated mutations that result in activation of signaling and report new inhibitory antibodies. We determined the structural basis for receptor inhibition by obtaining the first co-crystal structure of a NOTCH3 antibody with the NRR protein and defined two distinct epitopes for NRR antibodies. The antibodies exhibit potent anti-leukemic activity in cell lines and tumor xenografts harboring NOTCH3 activating mutations. Screening of primary T-ALL samples reveals that 2 of 40 tumors examined show active NOTCH3 signaling. We also identified evidence of NOTCH3 activation in 12 of 24 patient-derived orthotopic xenograft models, 2 of which exhibit activation of NOTCH3 without activation of NOTCH1. Our studies provide additional insights into NOTCH3 activation and offer a path forward for identification of cancers that are likely to respond to therapy with NOTCH3 selective inhibitory antibodies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kopan R, Ilagan MX . The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009; 137: 216–233.
Guruharsha KG, Kankel MW, Artavanis-Tsakonas S . The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 2012; 13: 654–666.
Andersson ER, Sandberg R, Lendahl U . Notch signaling: simplicity in design, versatility in function. Development 2011; 138: 3593–3612.
Fryer CJ, White JB, Jones KA . Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 2004; 16: 509–520.
Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood 2009; 113: 4381–4390.
Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol 2006; 26: 4642–4651.
Ntziachristos P, Lim JS, Sage J, Aifantis I . From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 2014; 25: 318–334.
Ranganathan P, Weaver KL, Capobianco AJ . Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer 2011; 11: 338–351.
Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 2011; 17: 1646–1651.
Stoeck A, Lejnine S, Truong A, Pan L, Wang H, Zang C et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov 2014; 4: 1154–1167.
Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest 2013; 123: 2965–2968.
Ho AS, Kannan K, Roy DM, Morris LG, Ganly I, Katabi N et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet 2013; 45: 791–798.
Nakayama K, Nakayama N, Jinawath N, Salani R, Kurman RJ, Shih Ie M et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer 2007; 120: 2613–2617.
Etemadmoghadam D, deFazio A, Beroukhim R, Mermel C, George J, Getz G et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res 2009; 15: 1417–1427.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615.
Aster JC, Blacklow SC . Targeting the Notch pathway: twists and turns on the road to rational therapeutics. J Clin Oncol 2012; 30: 2418–2420.
Vooijs M, Liu Z, Kopan R . Notch: architect, landscaper, and guardian of the intestine. Gastroenterology 2011; 141: 448–459.
Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev 2000; 14: 1343–1352.
Huppert SS, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 2000; 405: 966–970.
Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK . Essential role of endothelial Notch1 in angiogenesis. Circulation 2005; 111: 1826–1832.
McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M et al. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 2001; 128: 491–502.
Krebs LT, Xue Y, Norton CR, Sundberg JP, Beatus P, Lendahl U et al. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 2003; 37: 139–143.
Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev 2004; 18: 2730–2735.
Xu X, Choi SH, Hu T, Tiyanont K, Habets R, Groot AJ et al. Insights into autoregulation of Notch3 from structural and functional studies of its negative regulatory region. Structure 2015; 23: 1227–1235.
Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PloS One 2010; 5: e9094.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607.
Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol 2009; 27: 1025–1031.
Wang Z, Gerstein M, Snyder M . RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63.
Prassler J, Steidl S, Urlinger S . In vitro affinity maturation of HuCAL antibodies: complementarity determining region exchange and RapMAT technology. Immunotherapy 2009; 1: 571–583.
Prassler J, Thiel S, Pracht C, Polzer A, Peters S, Bauer M et al. HuCAL PLATINUM, a synthetic Fab library optimized for sequence diversity and superior performance in mammalian expression systems. J Mol Biol 2011; 413: 261–278.
Rauchenberger R, Borges E, Thomassen-Wolf E, Rom E, Adar R, Yaniv Y et al. Human combinatorial Fab library yielding specific and functional antibodies against the human fibroblast growth factor receptor 3. J Biol Chem 2003; 278: 38194–38205.
Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem 2008; 283: 8046–8054.
Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res 2008; 68: 1881–1888.
Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PloS One 2009; 4: e6613.
Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y et al. Therapeutic antibody targeting of individual Notch receptors. Nature 2010; 464: 1052–1057.
Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC . Structural basis for autoinhibition of Notch. Nat Struct Mol Biol 2007; 14: 295–300.
Chiang MY, Xu L, Shestova O, Histen G, L'Heureux S, Romany C et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 2008; 118: 3181–3194.
Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J 2000; 19: 3337–3348.
Tiyanont K, Wales TE, Siebel CW, Engen JR, Blacklow SC . Insights into Notch3 activation and inhibition mediated by antibodies directed against its negative regulatory region. J Mol Biol 2013; 425: 3192–3204.
Wang K, Zhang Q, Li D, Ching K, Zhang C, Zheng X et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a gamma-secretase inhibitor. Clin Cancer Res 2015; 21: 1487–1496.
Kluk MJ, Ashworth T, Wang H, Knoechel B, Mason EF, Morgan EA et al. Gauging NOTCH1 activation in cancer using immunohistochemistry. PloS One 2013; 8: e67306.
Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003; 425: 841–846.
Ramasamy SK, Kusumbe AP, Wang L, Adams RH . Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 2014; 507: 376–380.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Berman H, Henrick K, Nakamura H . Announcing the worldwide protein data bank. Nat Struct Biol 2003; 10: 980.
Acknowledgements
The JAGGED1 and DLL1 expressing cell lines were kindly provide by Dr Gerry Weinmaster (UCLA). HPB-ALL cells were kindly provided by Andreas Stasser (Walter and Eliza Hall Institute for Medical Research, Australia). We thank the Novartis Biologics Center including Thomas Pietzonka, Janine Shulok, Nadine Charara, Bill Tschantz and Tony Fleming. We also thank many senior Novartis leaders for supporting this project, including Jeff Porter and William Sellers. We thank Rajiv Chopra and Kirk Clark of Novartis Institutes for BioMedical Research for the instructive discussion on crystallography experiments. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman–Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was financially supported in part by the Dana Farber–Novartis DDP, and grants from the National Institutes of Health (P01 CA119070) and the Leukemia and Lymphoma Society (7003-13).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
P Bernasconi-Elias, T Hu, B Firestone, S Gans, E Kurth, P Capodieci, P LeMotte, A London, E Nolin, M Jones, K Slocum, are employees of Novartis Institutes for Biomedical Research. J Deplazes-Lauber, K Petropoulos, J Jaehrling are employees of MorphoSys AG. S Blacklow serves as a consultants for Novartis and receives research support through the Dana Farber–Novartis DDP. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Bernasconi-Elias, P., Hu, T., Jenkins, D. et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene 35, 6077–6086 (2016). https://doi.org/10.1038/onc.2016.133
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.133
This article is cited by
-
Epigenetically dysregulated NOTCH-Delta-HES signaling cascade can serve as a subtype classifier for acute lymphoblastic leukemia
Annals of Hematology (2024)
-
BGB-A445, a novel non-ligand-blocking agonistic anti-OX40 antibody, exhibits superior immune activation and antitumor effects in preclinical models
Frontiers of Medicine (2023)
-
Notch signaling pathway: architecture, disease, and therapeutics
Signal Transduction and Targeted Therapy (2022)
-
The proprotein convertase furin in cancer: more than an oncogene
Oncogene (2022)
-
Targeting Notch in oncology: the path forward
Nature Reviews Drug Discovery (2021)