Abstract
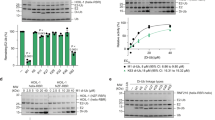
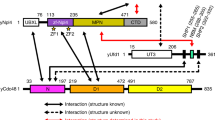
OTUB1 is a Lys48-specific deubiquitinating enzyme that forms a complex in vivo with E2 ubiquitin (Ub)-conjugating enzymes including UBC13 and UBCH5. OTUB1 binds E2~Ub thioester intermediates and prevents ubiquitin transfer, thereby noncatalytically inhibiting accumulation of polyubiquitin. We report here that a second role of OTUB1-E2 interactions is to stimulate OTUB1 cleavage of Lys48 polyubiquitin. This stimulation is regulated by the ratio of charged to uncharged E2 and by the concentration of Lys48-linked polyubiquitin and free ubiquitin. Structural and biochemical studies of human and worm OTUB1 and UBCH5B show that the E2 enzyme stimulates binding of the Lys48 polyubiquitin substrate by stabilizing folding of the OTUB1 N-terminal ubiquitin-binding helix. Our results suggest that OTUB1–E2 complexes in the cell are poised to regulate polyubiquitin chain elongation or degradation in response to changing levels of E2 charging and available free ubiquitin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Komander, D., Clague, M.J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Amerik, A.Y., Swaminathan, S., Krantz, B.A., Wilkinson, K.D. & Hochstrasser, M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16, 4826–4838 (1997).
Balakirev, M.Y., Tcherniuk, S.O., Jaquinod, M. & Chroboczek, J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4, 517–522 (2003).
Wang, T. et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J. Mol. Biol. 386, 1011–1023 (2009).
Soares, L. et al. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5, 45–54 (2004).
Li, S. et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285, 4291–4297 (2010).
Sun, X.X., Challagundla, K.B. & Dai, M.S. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 31, 576–592 (2012).
Goncharov, T. et al. OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 32, 1103–1114 (2013).
Nakada, S. et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010).
Sowa, M.E., Bennett, E.J., Gygi, S.P. & Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009).
Wiener, R., Zhang, X., Wang, T. & Wolberger, C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 (2012).
Juang, Y.C. et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 (2012).
Edelmann, M.J. et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390 (2009).
Jin, J., Li, X., Gygi, S.P. & Harper, J.W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007).
Haas, A.L. & Bright, P.M. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J. Biol. Chem. 262, 345–351 (1987).
Plechanovová, A. et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18, 1052–1059 (2011).
Pruneda, J.N., Stoll, K.E., Bolton, L.J., Brzovic, P.S. & Klevit, R.E. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry 50, 1624–1633 (2011).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Ventii, K.H. & Wilkinson, K.D. Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 (2008).
Wing, S.S. Deubiquitinating enzymes: the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell Biol. 35, 590–605 (2003).
Dantuma, N.P., Groothuis, T.A., Salomons, F.A. & Neefjes, J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173, 19–26 (2006).
Kimura, Y. et al. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell 137, 549–559 (2009).
Stanisić, V., Malovannaya, A., Qin, J., Lonard, D.M. & O'Malley, B.W. OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) α and affects ERα transcriptional activity. J. Biol. Chem. 284, 16135–16145 (2009).
Plechanovová, A., Jaffray, E., Tatham, M.H., Naismith, J.H. & Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Reverter, D. & Lima, C.D. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12, 1519–1531 (2004).
Sakata, E. et al. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure 18, 138–147 (2010).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Ozkan, E., Yu, H. & Deisenhofer, J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 102, 18890–18895 (2005).
Vijay-Kumar, S., Bugg, C.E. & Cook, W.J. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194, 531–544 (1987).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Winn, M.D., Murshudov, G.N. & Papiz, M.Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003).
Afonine, P.V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We thank S. Urbé and J.W. Harper for advice and suggestions. This work was supported in part by US National Institutes of Health grant GM-060980 (M.J.M.) and National Science Foundation grant MCB-0920082 (C.W.).
Author information
Authors and Affiliations
Contributions
R.W., A.T.D. and C.W. designed experiments, interpreted data and wrote the paper. R.W. and A.T.D. carried out the enzymatic assays and binding studies, and A.T.D. determined the crystal structure and did western blot assays. X.Z. purified proteins and prepared and crystallized the OTUB1–Ubal–UBCH5B~Ub complex. P.M.L. assayed E2–OTUB1 activity on different polyubiquitin chains. C.M.G. performed cell-based experiments under the guidance of M.J.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 7503 kb)
Rights and permissions
About this article
Cite this article
Wiener, R., DiBello, A., Lombardi, P. et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol 20, 1033–1039 (2013). https://doi.org/10.1038/nsmb.2655
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2655
This article is cited by
-
Deubiquitinases in cancer
Nature Reviews Cancer (2023)
-
The Role of Ubiquitin–Proteasome System and Mitophagy in the Pathogenesis of Parkinson's Disease
NeuroMolecular Medicine (2023)
-
Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nature Communications (2022)
-
Inhibition of cathepsin K sensitizes oxaliplatin-induced apoptotic cell death by Bax upregulation through OTUB1-mediated p53 stabilization in vitro and in vivo
Oncogene (2022)
-
Deubiquitinase-targeting chimeras for targeted protein stabilization
Nature Chemical Biology (2022)