Abstract
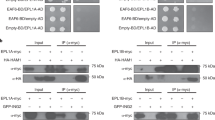
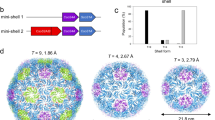
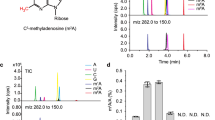
Chromodomains typically recruit protein complexes to chromatin and read the epigenetic histone code by recognizing lysine methylation in histone tails. We report the crystal structure of the chloroplast signal recognition particle (cpSRP) core from Arabidopsis thaliana, with the cpSRP54 tail comprising an arginine-rich motif bound to the second chromodomain of cpSRP43. A twinned aromatic cage reads out two neighboring nonmethylated arginines and adapts chromodomains to a non-nuclear function in post-translational targeting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Brehm, A., Tufteland, K.R., Aasland, R. & Becker, P.B. Bioessays 26, 133–140 (2004).
Paro, R. & Hogness, D.S. Proc. Natl. Acad. Sci. USA 88, 263–267 (1991).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Bannister, A.J. et al. Nature 410, 120–124 (2001).
Andersen, C.B. et al. Nature 443, 663–668 (2006).
Richter, C.V., Bals, T. & Schunemann, D. Eur. J. Cell Biol. 89, 965–973 (2010).
Klimyuk, V.I. et al. Plant Cell 11, 87–99 (1999).
Stengel, K.F. et al. Science 321, 253–256 (2008).
Grudnik, P., Bange, G. & Sinning, I. Biol. Chem. 390, 775–782 (2009).
Schuenemann, D. et al. Proc. Natl. Acad. Sci. USA 95, 10312–10316 (1998).
Falk, S. & Sinning, I. J. Biol. Chem. 285, 21655–21661 (2010).
Goforth, R.L. et al. J. Biol. Chem. 279, 43077–43084 (2004).
Falk, S., Ravaud, S., Koch, J. & Sinning, I. J. Biol. Chem. 285, 5954–5962 (2010).
Funke, S., Knechten, T., Ollesch, J. & Schunemann, D. J. Biol. Chem. 280, 8912–8917 (2005).
Jacobs, S.A. & Khorasanizadeh, S. Science 295, 2080–2083 (2002).
Fischle, W. et al. Genes Dev. 17, 1870–1881 (2003).
Hughes, R.M., Wiggins, K.R., Khorasanizadeh, S. & Waters, M.L. Proc. Natl. Acad. Sci. USA 104, 11184–11188 (2007).
Sivaraja, V. et al. J. Biol. Chem. 280, 41465–41471 (2005).
Gorinsek, B., Gubensek, F. & Kordis, D. Cytogenet. Genome Res. 110, 543–552 (2005).
Rosenblad, M.A., Larsen, N., Samuelsson, T. & Zwieb, C. RNA Biol. 6, 508–516 (2009).
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) SFB638 and the Graduiertenkolleg GRK1188 (to I.S.). We thank J. Kopp and C. Siegmann from the crystallization platform of the Biochemiezentrum der Universität Heidelberg (BZH)/Cluster of Excellence:CellNetworks for support in protein crystallization, R. Lindner for support in evolutionary analysis, and S. Falk, G. Bange and D. Schünemann for stimulating discussions. We acknowledge the European Synchrotron Radiation Facility for providing synchrotron radiation and for assistance in using beamlines ID14eh2 and ID14eh3. M.S. and H.M. acknowledge support from the Cluster of Excellence, Center for integrated Protein Science, Munich (CiPSM). I.S. is an investigator of the Cluster of Excellence:CellNetworks.
Author information
Authors and Affiliations
Contributions
I.H., K.W. and I.S. designed experiments and analyzed the data. I.H. conducted experiments. N.H.M. and M.S. provided NMR spectroscopy data and analysis. A.R. processed and analyzed SAXS data. I.H., K.W. and I.S. wrote the manuscript. All authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 10415 kb)
Rights and permissions
About this article
Cite this article
Holdermann, I., Meyer, N., Round, A. et al. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol 19, 260–263 (2012). https://doi.org/10.1038/nsmb.2196
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2196
This article is cited by
-
Chloroplast SRP43 autonomously protects chlorophyll biosynthesis proteins against heat shock
Nature Plants (2021)
-
Molecular mechanism of SRP-dependent light-harvesting protein transport to the thylakoid membrane in plants
Photosynthesis Research (2018)
-
Complementation of a mutation in CpSRP43 causing partial truncation of light-harvesting chlorophyll antenna in Chlorella vulgaris
Scientific Reports (2017)
-
Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction
Nature Communications (2015)