Abstract
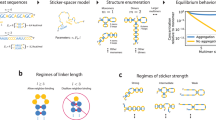
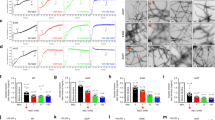
Because polyglutamine (polyQ) aggregate formation has been implicated as playing an important role in expanded CAG repeat diseases, it is important to understand the biophysics underlying the initiation of aggregation. Previously, we showed that relatively long polyQ peptides aggregate by nucleated growth polymerization and a monomeric critical nucleus. We show here that over a short range of repeat lengths, from Q23 to Q26, the size of the critical nucleus for aggregation increases from monomeric to dimeric to tetrameric. This variation in nucleus size suggests a common duplex antiparallel β-sheet framework for the nucleus, and it further supports the feasibility of an organized monomeric aggregation nucleus for longer polyQ repeat peptides. The data also suggest that a change in the size of aggregation nuclei may have a role in the pathogenicity of polyQ expansion in this series of familial neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
References
Bates, G.P. & Benn, C. The polyglutamine diseases. in Huntington's Disease (eds. Bates, G.P., Harper, P.S. & Jones, L.) 429–472 (Oxford University Press, Oxford, UK, 2002).
Ross, C.A. & Poirier, M.A. Opinion: What is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 6, 891–898 (2005).
Scherzinger, E. et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 (1997).
Chen, S., Berthelier, V., Yang, W. & Wetzel, R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J. Mol. Biol. 311, 173–182 (2001).
Davies, S.W. et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 (1997).
Chen, S., Ferrone, F. & Wetzel, R. Huntington's disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. USA 99, 11884–11889 (2002).
Bhattacharyya, A.M., Thakur, A.K. & Wetzel, R. Polyglutamine aggregation nucleation: thermodynamics of a highly unfavorable protein folding reaction. Proc. Natl. Acad. Sci. USA 102, 15400–15405 (2005).
Poirier, M.A. et al. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J. Biol. Chem. 277, 41032–41037 (2002).
Wacker, J.L., Zareie, M.H., Fong, H., Sarikaya, M. & Muchowski, P.J. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 11, 1215–1222 (2004).
Thakur, A.K. et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 16, 380–389 (2009).
Bulone, D., Masino, L., Thomas, D.J., San Biagio, P.L. & Pastore, A. The interplay between PolyQ and protein context delays aggregation by forming a reservoir of protofibrils. PLoS One 1, e111 (2006).
Ignatova, Z., Thakur, A.K., Wetzel, R. & Gierasch, L.M. In-cell aggregation of a polyglutamine-containing chimera is a multistep process initiated by the flanking sequence. J. Biol. Chem. 282, 36736–36743 (2007).
de Chiara, C., Menon, R.P., Dal Piaz, F., Calder, L. & Pastore, A. Polyglutamine is not all: the functional role of the AXH domain in the ataxin-1 protein. J. Mol. Biol. 354, 883–893 (2005).
Ellisdon, A.M., Thomas, B. & Bottomley, S.P. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J. Biol. Chem. 281, 16888–16896 (2006).
Bhattacharyya, A. et al. Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 355, 524–535 (2006).
Gu, X. et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron 64, 828–840 (2009).
Romero, P., Obradovic, Z. & Dunker, A.K. Natively disordered proteins: functions and predictions. Appl. Bioinformatics 3, 105–113 (2004).
Mohan, A. et al. Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 (2006).
Ferrone, F. Analysis of protein aggregation kinetics. Methods Enzymol. 309, 256–274 (1999).
Thakur, A.K. & Wetzel, R. Mutational analysis of the structural organization of polyglutamine aggregates. Proc. Natl. Acad. Sci. USA 99, 17014–17019 (2002).
Slepko, N. et al. Normal-repeat-length polyglutamine peptides accelerate aggregation nucleation and cytotoxicity of expanded polyglutamine proteins. Proc. Natl. Acad. Sci. USA 103, 14367–14372 (2006).
Jayaraman, M., Kodali, R. & Wetzel, R. The impact of ataxin-1-like histidine insertions on polyglutamine aggregation. Protein Eng. Des. Sel. 22, 469–478 (2009).
Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N. & Goldberg, A.L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them intact during degradation of polyglutamine-containing proteins. Mol. Cell 14, 95–104 (2004).
Kirkitadze, M.D., Condron, M.M. & Teplow, D.B. Identification and characterization of key kinetic intermediates in amyloid beta-protein fibrillogenesis. J. Mol. Biol. 312, 1103–1119 (2001).
Lee, C.C., Walters, R.H. & Murphy, R.M. Reconsidering the mechanism of polyglutamine peptide aggregation. Biochemistry 46, 12810–12820 (2007).
Chen, S. & Wetzel, R. Solubilization and disaggregation of polyglutamine peptides. Protein Sci. 10, 887–891 (2001).
Wetzel, R. Protein folding and aggregation in the expanded polyglutamine repeat diseases. in The Protein Folding Handbook (eds Buchner, J. & Kiefhaber, T.) Part II, 1170–1214 (Wiley-VCH, Weinheim, 2005).
O'Nuallain, B. et al. Kinetics and thermodynamics of amyloid assembly using a high-performance liquid chromatography-based sedimentation assay. Methods Enzymol. 413, 34–74 (2006).
Legleiter, J. et al. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J. Biol. Chem. 285, 14777–14790 (2010).
Heller, J. et al. Solid-state NMR studies of the prion protein H1 fragment. Protein Sci. 5, 1655–1661 (1996).
Chen, S., Berthelier, V., Hamilton, J.B., O'Nuallain, B. & Wetzel, R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 41, 7391–7399 (2002).
Jayaraman, M., Thakur, A.K., Kar, K., Kodali, R. & Wetzel, R. Assays for studying nucleated aggregation of polyglutamine proteins. Methods published online, doi:10.1016/j.ymeth.2011.01.001 (11 January 2011).
Frieden, C. Protein aggregation processes: in search of the mechanism. Protein Sci. 16, 2334–2344 (2007).
O'Nuallain, B., Shivaprasad, S., Kheterpal, I. & Wetzel, R. Thermodynamics of Aβ(1–40) amyloid fibril elongation. Biochemistry 44, 12709–12718 (2005).
Cannon, M.J., Williams, A.D., Wetzel, R. & Myszka, D.G. Kinetic analysis of β-amyloid fibril elongation. Anal. Biochem. 328, 67–75 (2004).
Ferrone, F.A. Nucleation: the connections between equilibrium and kinetic behavior. Methods Enzymol. 412, 285–299 (2006).
Tam, S. et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16, 1279–1285 (2009).
Arvinte, T. Human calcitonin fibrillogenesis. in Ciba Foundation Symp. 199—The Nature and Origin of Amyloid Fibrils (ed. Bock, G.R.) 90–97 (Wiley, New York, 1996).
Collins, S.R., Douglass, A., Vale, R.D. & Weissman, J.S. Mechanism of prion propagation: amyloid growth occurs by monomer addition. PLoS Biol. 2, e321 (2004).
Frankenfield, K.N., Powers, E.T. & Kelly, J.W. Influence of the N-terminal domain on the aggregation properties of the prion protein. Protein Sci. 14, 2154–2166 (2005).
Xue, W.F., Homans, S.W. & Radford, S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA 105, 8926–8931 (2008).
Walters, R.H. & Murphy, R.M. Examining polyglutamine peptide length: a connection between collapsed conformations and increased aggregation. J. Mol. Biol. 393, 978–992 (2009).
Khare, S.D., Ding, F., Gwanmesia, K.N. & Dokholyan, N.V. Molecular origin of polyglutamine aggregation in neurodegenerative diseases. PLOS Comput. Biol. 1, 230–235 (2005).
Marchut, A.J. & Hall, C.K. Effects of chain length on the aggregation of model polyglutamine peptides: molecular dynamics simulations. Proteins 66, 96–109 (2007).
Smith, M.H. et al. Polyglutamine fibrils are formed using a simple designed beta-hairpin model. Proteins 78, 1971–1979 (2010).
Sharma, D., Shinchuk, L.M., Inouye, H., Wetzel, R. & Kirschner, D.A. Polyglutamine homopolymers having 8–45 residues form slablike β-crystallite assemblies. Proteins 61, 398–411 (2005).
Sikorski, P. & Atkins, E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules 6, 425–432 (2005).
Crick, S.L., Jayaraman, M., Frieden, C., Wetzel, R. & Pappu, R.V. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc. Natl. Acad. Sci. USA 103, 16764–16769 (2006).
Dougan, L., Li, J.Y., Badilla, C.L., Berne, B.J. & Fernandez, J.M. Single homopolypeptide chains collapse into mechanically rigid conformations. Proc. Natl. Acad. Sci. USA 106, 12605–12610 (2009).
Wang, X., Vitalis, A., Wyczalkowski, M.A. & Pappu, R.V. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins 63, 297–311 (2006).
Starikov, E.B., Lehrach, H. & Wanker, E.E. Folding of oligoglutamines: a theoretical approach based upon thermodynamics and molecular mechanics. J. Biomol. Struct. Dyn. 17, 409–427 (1999).
Esposito, L., Paladino, A., Pedone, C. & Vitagliano, L. Insights into structure, stability, and toxicity of monomeric and aggregated polyglutamine models from molecular dynamics simulations. Biophys. J. 94, 4031–4040 (2008).
Lakhani, V.V., Ding, F. & Dokholyan, N.V. Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLOS Comput. Biol. 6, e1000772 (2010).
Nagai, Y. et al. A toxic monomeric conformer of the polyglutamine protein. Nat. Struct. Mol. Biol. 14, 332–340 (2007).
Acknowledgements
We acknowledge access to the PONDR® analysis, provided by Molecular Kinetics. We thank G. Calero for access to his dynamic light-scattering instrument. EMs were collected in the University of Pittsburgh School of Medicine Structural Biology Department's EM facility administered by J. Conway and A. Makhov. We gratefully acknowledge funding support from US National Institutes of Health grants R01 AG019322 and R21 AG033757 (R.W.).
Author information
Authors and Affiliations
Contributions
K.K., M.J. and B.S. purified the peptides, determined and analyzed the aggregation kinetics and obtained dynamic light-scattering data on aggregation time points. R.K. obtained the EM and FTIR data. R.W. wrote the paper. All authors contributed to study design, data interpretation and improving the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 565 kb)
Rights and permissions
About this article
Cite this article
Kar, K., Jayaraman, M., Sahoo, B. et al. Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat Struct Mol Biol 18, 328–336 (2011). https://doi.org/10.1038/nsmb.1992
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1992
This article is cited by
-
Molecular mechanisms of heterogeneous oligomerization of huntingtin proteins
Scientific Reports (2019)
-
Distinct thermodynamic signatures of oligomer generation in the aggregation of the amyloid-β peptide
Nature Chemistry (2018)
-
Rapid α-oligomer formation mediated by the Aβ C terminus initiates an amyloid assembly pathway
Nature Communications (2016)
-
Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies
Experimental & Molecular Medicine (2015)
-
Alternative splicing of Drosophila Nmnat functions as a switch to enhance neuroprotection under stress
Nature Communications (2015)