Abstract
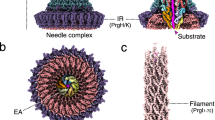
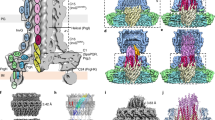
Pathogenic Gram-negative bacteria use a type three secretion system (TTSS) to deliver virulence factors into host cells. Although the order in which proteins incorporate into the growing TTSS is well described, the underlying assembly mechanisms are still unclear. Here we show that the TTSS needle protomer refolds spontaneously to extend the needle from the distal end. We developed a functional mutant of the needle protomer from Shigella flexneri and Salmonella typhimurium to study its assembly in vitro. We show that the protomer partially refolds from α-helix into β-strand conformation to form the TTSS needle. Reconstitution experiments show that needle growth does not require ATP. Thus, like the structurally related flagellar systems, the needle elongates by subunit polymerization at the distal end but requires protomer refolding. Our studies provide a starting point to understand the molecular assembly mechanisms and the structure of the TTSS at atomic level.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kubori, T. et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998).
Journet, L., Agrain, C., Broz, P. & Cornelis, G.R. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 (2003).
Tamano, K. et al. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19, 3876–3887 (2000).
Cornelis, G.R. The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 (2006).
Galan, J.E. & Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 (2006).
Nhieu, G.T. & Sansonetti, P.J. Mechanism of Shigella entry into epithelial cells. Curr. Opin. Microbiol. 2, 51–55 (1999).
Kimbrough, T.G. & Miller, S.I. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 4, 75–82 (2002).
Marlovits, T.C. et al. Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 (2004).
Blocker, A. et al. Structure and composition of the Shigella flexneri 'needle complex', a part of its type III secreton. Mol. Microbiol. 39, 652–663 (2001).
Blocker, A.J. et al. What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. USA 105, 6507–6513 (2008).
Kawagishi, I., Homma, M., Williams, A.W. & Macnab, R.M. Characterization of the flagellar hook length control protein fliK of Salmonella typhimurium and Escherichichia coli. J. Bacteriol. 178, 2954–2959 (1996).
Espina, M. et al. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect. Immun. 74, 4391–4400 (2006).
Mota, L.J. Type III secretion gets an LcrV tip. Trends Microbiol. 14, 197–200 (2006).
Mueller, C.A. et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676 (2005).
Sani, M. et al. IpaD is localized at the tip of the Shigella flexneri type III secretion apparatus. Biochim. Biophys. Acta 1770, 307–311 (2007).
Rogers, D.R. Screening for amyoloid with the thioflavin-T fluorescent method. Am. J. Clin. Pathol. 44, 59–61 (1965).
Blake, C.C.F. et al. Nature and Origin of Amyloid Fibrils. CIBA Foundation Symposium, No. 199 (1996).
Cordes, F.S. et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278, 17103–17107 (2003).
Quinaud, M. et al. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280, 36293–36300 (2005).
Marlovits, T.C. et al. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 (2006).
Bishop, M.F. & Ferrone, F.A. Kinetics of nucleation-controlled polymerization. A perturbation treatment for use with a secondary pathway. Biophys. J. 46, 631–644 (1984).
Emerson, S.U., Tokuyasu, K. & Simon, M.I. Bacterial flagella: polarity of elongation. Science 169, 190–192 (1970).
Wang, Y. et al. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371, 1304–1314 (2007).
Akeda, Y. & Galan, J.E. Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915 (2005).
Darboe, N., Kenjale, R., Picking, W.L., Picking, W.D. & Middaugh, C.R. Chemical denaturation of MxiH from Shigella and PrgI from Salmonella. Protein Sci. 15, 543–552 (2006).
Deane, J.E. et al. Expression, purification, crystallization and preliminary crystallographic analysis of MxiH, a subunit of the Shigella flexneri type III secretion system needle. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 62, 302–305 (2006).
Zhang, L., Wang, Y., Picking, W.L., Picking, W.D. & De Guzman, R.N. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 359, 322–330 (2006).
Barrett, B.S., Picking, W.L., Picking, W.D. & Middaugh, C.R. The response of type three secretion system needle proteins MxiHDelta5, BsaLDelta5, and PrgIDelta5 to temperature and pH. Proteins 73, 632–643 (2008).
Deane, J.E. et al. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl. Acad. Sci. USA 103, 12529–12533 (2006).
Kenjale, R. et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280, 42929–42937 (2005).
Oberg, K.A., Ruysschaert, J.M. & Goormaghtigh, E. Rationally selected basis proteins: a new approach to selecting proteins for spectroscopic secondary structure analysis. Protein Sci. 12, 2015–2031 (2003).
Quinaud, M. et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. USA 104, 7803–7808 (2007).
Sun, P., Tropea, J.E., Austin, B.P., Cherry, S. & Waugh, D.S. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377, 819–830 (2008).
Tan, Y.W., Yu, H.B., Leung, K.Y., Sivaraman, J. & Mok, Y.K. Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila. Protein Sci. 17, 1748–1760 (2008).
Minamino, T. & Namba, K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451, 485–488 (2008).
Paul, K., Erhardt, M., Hirano, T., Blair, D.F. & Hughes, K.T. Energy source of flagellar type III secretion. Nature 451, 489–492 (2008).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Padilla, J.E. & Yeates, T.O. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 59, 1124–1130 (2003).
Brunger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Kubori, T. et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998).
Delaglio, F. et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Keller, R. The Computer Aided Resonance Assignment Tutorial 1st edn. (CANTINA Verlag, Goldau, Switzerland, 2004).
Bodenhausen, G. & Ruben, D.J. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69, 185–189 (1980).
Grzesiek, S. & Bax, A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 3, 185–204 (1993).
Kay, L.E., Keifer, P. & Saarinen, T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 (1992).
Wittekind, M. & Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. B 101, 201–205 (1993).
Bax, A. & Ikura, M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the α-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J. Biomol. NMR 1, 99–104 (1991).
Grzesiek, S. et al. Proton, carbon-13, and nitrogen-15 NMR backbone assignments and secondary structure of human interferon-γ. Biochemistry 31, 8180–8190 (1992).
Ikura, M., Kay, L.E. & Bax, A. A novel approach for sequential assignment of proton, carbon-13, and nitrogen-15 spectra of larger proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29, 4659–4667 (1990).
Bax, A., Clore, G.M. & Gronenborn, A.M. 1H-1H correlation via isotropic mixing of carbon-13 magnetization, a new three-dimensional approach for assigning proton and carbon-13 spectra of carbon-13-enriched proteins. J. Magn. Reson. 88, 425–431 (1990).
Pervushin, K., Riek, R., Wider, G. & Wuthrich, K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA 94, 12366–12371 (1997).
Lee, D., Vogeli, B. & Pervushin, K. Detection of C′,Cα correlations in proteins using a new time- and sensitivity-optimal experiment. J. Biomol. NMR 31, 273–278 (2005).
Baldus, M. Molecular Interactions investigated by multi-dimensional solid-state NMR. Curr. Opin. Struct. Biol. 16, 618–623 (2006).
Seidel, K. et al. Protein solid-state NMR resonance assignments from (13C,13C) correlation spectroscopy. Phys. Chem. Chem. Phys. 6, 5090–5093 (2004).
Acknowledgements
We thank P. Jungblut and M. Schmid for help with MS analysis, V. Brinkmann and U. Abu Abed for help with electron microscopy sample preparation, U. Müller and S. Monaco for assistance in using beamlines, M. Luft for providing the opportunity of the FTIR measurements and for helpful discussions, B. Angerstein for help with ssNMR sample preparation, D. Lee for help in using Bruker spectrometers and B. Raupach, A. Zumsteg, F. Meissner, M. Lunelli and R. Kumar Lokareddy for their useful comments and critical reading of the manuscript. This work was supported by the Max-Planck-Society (to C. Griesinger and A.Z.) and by the Deutsche Forschungsgemeinschaft through an Emmy Noether stipend (LA 2705/1-1) to A.L.
Author information
Authors and Affiliations
Contributions
Ö.P. cloned constructs and purified protomer for every experiment, crystallized, collected, processed and refined X-ray diffraction data and performed cellular assays; H.S. collected, processed and analyzed liquid-state NMR data and performed FTIR experiments and ThioflavinT binding assays; F.D. performed and analyzed DLS and X-ray fiber diffraction experiments; K.S. and C.A. collected, processed and analyzed solid-state NMR data; H.T. and Ö.P. purified and performed in vitro growth experiments with TTSS; C. Goosmann performed TEM studies; A.L. and M.B. designed and analyzed solid-state NMR experiments; C. Griesinger designed and analyzed liquid-state NMR experiments; A.T. designed and analyzed DLS and X-ray fiber diffraction experiments; V.B. designed TEM experiments; A.Z. designed functional and structural experiments; M.K. conceived this study, designed functional and TEM experiments, collected, refined and analyzed X-ray diffraction data and wrote the paper; all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14, Supplementary Table 1 and Supplementary Methods (PDF 4245 kb)
Rights and permissions
About this article
Cite this article
Poyraz, Ö., Schmidt, H., Seidel, K. et al. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol 17, 788–792 (2010). https://doi.org/10.1038/nsmb.1822
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1822
This article is cited by
-
S-nitrosylation of UCHL1 induces its structural instability and promotes α-synuclein aggregation
Scientific Reports (2017)
-
Secretion systems in Gram-negative bacteria: structural and mechanistic insights
Nature Reviews Microbiology (2015)
-
Amino acids 89–96 of Salmonella typhimurium flagellin represent the major domain responsible for TLR5-independent adjuvanticity in the humoral immune response
Cellular & Molecular Immunology (2015)