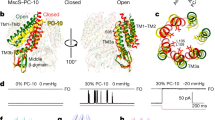
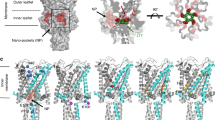
Abstract
Under prolonged stimulation, the mechanosensitive channel MscS of Escherichia coli enters a tension-insensitive inactivated state. We transformed the delipidated crystal structure and restored the link between lipid-facing TM1 and TM2 and gate-forming TM3 helices. Joining the conserved Phe68 of TM2 with Leu111 of TM1, this buried contact mediated opening in steered molecular dynamics simulations with forces applied to the peripheral helices. Both F68S and L111S substitutions produced severe loss-of-function phenotypes in vivo by increasing the inactivation rate and promoting unusual 'silent' inactivation from the resting state. F68S also suppressed the noninactivating phenotype of G113A. The L111C cysteine buried in the TM2-TM3 crevice was accessible to methanethiosulfonate-ethyltrimethylammonium (MTSET) only in the inactivated state, which was stabilized upon modification by a positive charge. The restored interhelical contact thus is critically involved in force transmission from the lipid-facing helices to the gate, and inactivation appears to be a result of TM2-TM3 uncoupling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Levina, N. et al. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18, 1730–1737 (1999).
Sukharev, S. Purification of the small mechanosensitive channel of Escherichia coli (MscS): the subunit structure, conduction, and gating characteristics in liposomes. Biophys. J. 83, 290–298 (2002).
Okada, K., Moe, P.C. & Blount, P. Functional design of bacterial mechanosensitive channels. Comparisons and contrasts illuminated by random mutagenesis. J. Biol. Chem. 277, 27682–27688 (2002).
Pivetti, C.D. et al. Two families of mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 67, 66–85 (2003).
Balleza, D. & Gomez-Lagunas, F. Conserved motifs in mechanosensitive channels MscL and MscS. Eur. Biophys. J. 38, 1013–1027 (2009).
Haswell, E.S. & Meyerowitz, E.M. MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16, 1–11 (2006).
Nakayama, Y., Fujiu, K., Sokabe, M. & Yoshimura, K. Molecular and electrophysiological characterization of a mechanosensitive channel expressed in the chloroplasts of Chlamydomonas. Proc. Natl. Acad. Sci. USA 104, 5883–5888 (2007).
Rayavara, K. & Desai, S.A. Genetic disruption of a mechanosensitive ion channel in Plasmodium falciparum. Am. J. Trop. Med. Hyg. 79, 65–66 (2008).
Akitake, B., Anishkin, A. & Sukharev, S. The “dashpot” mechanism of stretch-dependent gating in MscS. J. Gen. Physiol. 125, 143–154 (2005).
Grajkowski, W., Kubalski, A. & Koprowski, P. Surface changes of the mechanosensitive channel MscS upon its activation, inactivation, and closing. Biophys. J. 88, 3050–3059 (2005).
Bass, R.B., Strop, P., Barclay, M. & Rees, D.C. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587 (2002).
Steinbacher, S., Bass, R., Strop, P. & Rees, D.C. Structures of the prokaryotic mechanosensitive channels MscL and MscS. in Mechanosensitive Ion Channels (ed. Hamill, P.O.) Ch. 1 (Academic Press, 2007).
Wang, W. et al. The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science 321, 1179–1183 (2008).
Vasquez, V., Sotomayor, M., Cordero-Morales, J., Schulten, K. & Perozo, E. A structural mechanism for MscS gating in lipid bilayers. Science 321, 1210–1214 (2008).
Vasquez, V. et al. Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 378, 55–70 (2008).
Akitake, B., Anishkin, A., Liu, N. & Sukharev, S. Straightening and sequential buckling of the pore-lining helices define the gating cycle of MscS. Nat. Struct. Mol. Biol. 14, 1141–1149 (2007).
Anishkin, A. & Sukharev, S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys. J. 86, 2883–2895 (2004).
Sotomayor, M. & Schulten, K. Molecular dynamics study of gating in the mechanosensitive channel of small conductance MscS. Biophys. J. 87, 3050–3065 (2004).
Spronk, S.A., Elmore, D.E. & Dougherty, D.A. Voltage-dependent hydration and conduction properties of the hydrophobic pore of the mechanosensitive channel of small conductance. Biophys. J. 90, 3555–3569 (2006).
Nomura, T., Sokabe, M. & Yoshimura, K. Interaction between the cytoplasmic and transmembrane domains of the mechanosensitive channel MscS. Biophys. J. 94, 1638–1645 (2008).
Anishkin, A., Akitake, B. & Sukharev, S. Characterization of the resting MscS: modeling and analysis of the closed bacterial mechanosensitive channel of small conductance. Biophys. J. 94, 1252–1266 (2008).
Anishkin, A., Kamaraju, K. & Sukharev, S. Mechanosensitive channel MscS in the open state: modeling of the transition, explicit simulations, and experimental measurements of conductance. J. Gen. Physiol. 132, 67–83 (2008).
Eisenberg, D., Schwarz, E., Komaromy, M. & Wall, R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 (1984).
Kullback, S. Information Theory and Statistics (Dover Publications, Inc., Mineola, New York, USA, 1997).
Beckstein, O. & Sansom, M.S. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys. Biol. 1, 42–52 (2004).
Booth, I.R. et al. Structure-function relations of MscS. in Mechanosensitive Ion Channels (ed. Hamill, P.O.) Ch. 10 (Academic Press, 2007).
Sukharev, S., Akitake, B. & Anishkin, A. The bacterial mechanosensitive channel MscS: emerging principles of gating and modulation. in Mechanosensitive Ion Channels (ed. Hamill, P.O.) Ch. 9 (Academic Press, 2007).
Nomura, T., Sokabe, M. & Yoshimura, K. Lipid-protein interaction of the MscS mechanosensitive channel examined by scanning mutagenesis. Biophys. J. 91, 2874–2881 (2006).
White, S.H. & Wimley, W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 (1999).
Zhou, X. et al. Yeast screens show aromatic residues at the end of the sixth helix anchor transient receptor potential channel gate. Proc. Natl. Acad. Sci. USA 104, 15555–15559 (2007).
Su, Z. et al. Yeast gain-of-function mutations reveal structure-function relationships conserved among different subfamilies of transient receptor potential channels. Proc. Natl. Acad. Sci. USA 104, 19607–19612 (2007).
Chiang, C.S., Anishkin, A. & Sukharev, S. Gating of the large mechanosensitive channel in situ: estimation of the spatial scale of the transition from channel population responses. Biophys. J. 86, 2846–2861 (2004).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
MacKerell, A.D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald - an n·log(n) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Jorgensen, W.L., Chandrasekhar, J. & Madura, J.D. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 (1983).
Acknowledgements
The authors would like to thank M. Kiyatkin for compiling a library of MscS and MscK homologs and conducting the multiple sequence alignments, M. Boer for editing the text and I. Booth (Univ. of Aberdeen) and P. Blount (Univ. of Texas, Dallas) for kindly providing the MJF465 and PB113 strains, respectively. This work was supported by the US National Institutes of Health and an undergraduate research grant to V.B. from the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
V.B., A.A., K.K. and S.S. designed the experiments and analyzed the models and data; V.B. and K.K. conducted all of the patch-clamp experiments and generated some of the MscS mutants; A.A. performed all of the computations (extrapolated motion analysis and molecular dynamics simulations); N.L. generated all MscS mutants and prepared spheroplasts; V.B., A.A. and S.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Discussion (PDF 593 kb)
Supplementary Video 1
Simulated MscS opening (AVI 10111 kb)
Supplementary Data 1
MscS alignment, gap-free (TXT 36 kb)
Supplementary Data 2
MscS alignment, 86 Gram-negative species (TXT 28 kb)
Rights and permissions
About this article
Cite this article
Belyy, V., Anishkin, A., Kamaraju, K. et al. The tension-transmitting 'clutch' in the mechanosensitive channel MscS. Nat Struct Mol Biol 17, 451–458 (2010). https://doi.org/10.1038/nsmb.1775
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1775
This article is cited by
-
Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance
Nature Communications (2020)
-
Evolutionary specialization of MscCG, an MscS-like mechanosensitive channel, in amino acid transport in Corynebacterium glutamicum
Scientific Reports (2018)
-
Spatiotemporal relationships defining the adaptive gating of the bacterial mechanosensitive channel MscS
European Biophysics Journal (2018)
-
A single mechanism driving both inactivation and adaptation in rapidly adapting currents of DRG neurons?
Biological Cybernetics (2016)
-
On the gating of mechanosensitive channels by fluid shear stress
Acta Mechanica Sinica (2016)