Abstract
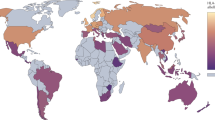
Multiple sclerosis (MS) is a common and severe CNS disorder that is characterized by myelin loss, chronic inflammation, axonal and oligodendrocyte pathology, and progressive neurological dysfunction. Extensive epidemiological data confirm that genetic variation is an important determinant of susceptibility to MS, and suggest that such variation also influences the timing of symptom onset, the course of the disease, and the treatment response. Multicenter international collaborations have allowed large and well-characterized sample collections to be assembled that, when coupled with high-powered laboratory technologies, afford the opportunity to analyze the genome with increasing resolution and detail. The seven MS genome-wide association screens that have been completed in the past 3 years have substantially lengthened the list of MS genetic risk associations. Nevertheless, our knowledge of MS genetics remains incomplete, with many risk alleles still to be revealed, although progress is likely to be rapid in the near future. The ensuing challenge will be to design effective functional studies that convincingly link genetic variation to the underlying pathophysiology of MS. Establishment of such connections might translate into clinically useful genetic biomarkers and reveal novel targets for therapy. This Review briefly summarizes well-established concepts of MS epidemiology and susceptibility, and discusses new knowledge emerging from genome-wide association studies.
Key Points
-
Genetic variation is an important determinant of susceptibility to and progression of multiple sclerosis (MS)
-
MS is one of the so-called complex genetic diseases, which are common disorders that are characterized by modest disease risk heritability and multifaceted gene–environment interactions
-
The human leukocyte antigen gene cluster represents by far the strongest MS susceptibility locus, and was identified in both candidate gene association and linkage studies
-
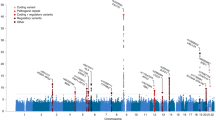
Genome-wide association studies have dramatically increased the number of MS risk associations; however, only a fraction of the heritability of this disease has been explained
-
With the aid of high-capacity technologies, next-generation studies will fully define the genetic mechanisms operating in MS and, hence, will assist in the formulation of a reliable model of pathogenesis
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pugliatti, M. et al. The epidemiology of multiple sclerosis in Europe. Eur. J. Neurol. 13, 700–722 (2006).
Hauser, S. L. & Oksenberg, J. R. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 52, 61–76 (2006).
Hauser, S. L. & Goodin, D. S. In Harrison's Principle of Internal Medicine 17th edn Ch. 375 (eds Fauci, A. D. et al.) 2611–2621 (McGraw Hill, New York, 2008).
Henderson, A. P., Barnett, M. H., Parratt, J. D. & Prineas, J. W. Multiple sclerosis: distribution of inflammatory cells in newly forming lesions. Ann. Neurol. 66, 739–753 (2009).
Lucchinetti, C. Pathological heterogeneity of idiopathic central nervous system inflammatory demyelinating disorders. Curr. Top. Microbiol. Immunol. 318, 19–43 (2008).
Breij, E. C. et al. Homogeneity of active demyelinating lesions in established multiple sclerosis. Ann. Neurol. 63, 16–25 (2008).
Alter, M., Leibowitz, U. & Speer, J. Risk of multiple sclerosis related to age at migration to Israel. Arch. Neurol. 15, 234–237 (1996).
Dean, G. & Kurtzke, J. F. On the risk of multiple sclerosis according to age at migration to South Africa. Br. Med. J. 3, 725–729 (1971).
Cabre, P. et al. Role of return migration in the emergence of multiple sclerosis in the French West Indies. Brain 128, 2899–2910 (2005).
Islam, T. et al. Differential twin concordance for multiple sclerosis by latitude of birthplace. Ann. Neurol. 60, 56–64 (2006).
Islam, T., Gauderman, W. J., Cozen, W. & Mack, T. M. Childhood sun exposure influences risk of multiple sclerosis in monozygotic twins. Neurology 69, 381–388 (2007).
Beretich, B. D. & Beretich, T. M. Explaining multiple sclerosis prevalence by ultraviolet exposure: a geospatial analysis. Mult. Scler. 15, 891–898 (2009).
Willer, C. J. et al. Timing of birth and risk of multiple sclerosis: population based study. BMJ 330, 120 (2005).
Sotgiu, S. et al. Seasonal fluctuation of multiple sclerosis births in Sardinia. J. Neurol. 253, 38–44 (2006).
Ramagopalan, S. V. et al. HLA-DRB1 and month of birth in multiple sclerosis. Neurology 73, 2107–2111 (2009).
Orton, S. M. et al. Sex ratio of multiple sclerosis in Canada: a longitudinal study. Lancet Neurol. 5, 932–936 (2006).
Kurtzke, J. F. Multiple sclerosis in time and space—geographic clues to cause. J. Neurovirol. 6 (Suppl. 2), S134–S140 (2000).
Pugliatti, M. et al. Evidence of early childhood as the susceptibility period in multiple sclerosis: space–time cluster analysis in a Sardinian population. Am. J. Epidemiol. 164, 326–333 (2006).
Oksenberg, J. R., Baranzini, S. E., Sawcer, S. & Hauser, S. L. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat. Rev. Genet. 9, 516–526 (2008).
Mackay, R. P. Familial occurrence of multiple sclerosis and its implications. Arch. Neurol. Psychiatry 64, 155–157 (1950).
Schapira, K., Poskanzer, D. C. & Miller, H. Familial and conjugal multiple sclerosis. Brain 86, 315–332 (1963).
Doolittle, T. H. et al. Multiple sclerosis sibling pairs: clustered onset and familial predisposition. Neurology 40, 1546–1552 (1990).
Robertson, N. P. et al. Age-adjusted recurrence risks for relatives of patients with multiple sclerosis. Brain 119, 449–455 (1996).
Risch, N. Linkage strategies for genetically complex traits. I. Multilocus models. Am. J. Hum. Genet. 46, 222–228 (1990).
Sadovnick, A. D. Familial recurrence risks and inheritance of multiple sclerosis. Curr. Opin. Neurol. Neurosurg. 6, 189–194 (1993).
Guo, S. W. Inflation of sibling recurrence-risk ratio, due to ascertainment bias and/or overreporting. Am. J. Hum. Genet. 63, 252–258 (1998).
Hemminki, K., Li, X., Sundquist, J., Hillert, J. & Sundquist, K. Risk for multiple sclerosis in relatives and spouses of patients diagnosed with autoimmune and related conditions. Neurogenetics 10, 5–11 (2009).
Sawcer, S., Ban, M., Wason, J. & Dudbridge, F. What role for genetics in the prediction of multiple sclerosis? Ann. Neurol. 67, 3–10 (2010).
Carton, H. et al. Risks of multiple sclerosis in relatives of patients in Flanders, Belgium. J. Neurol. Neurosurg. Psychiatry 62, 329–333 (1997).
Ebers, G. C., Sadovnick, A. D. & Risch, N. J. A genetic basis for familial aggregation in multiple sclerosis. Nature 377, 150–151 (1995).
Sadovnick, A. D., Ebers, G. C., Dyment, D. A. & Risch, N. J. Evidence for genetic basis of multiple sclerosis. The Canadian Collaborative Study Group. Lancet 347, 1728–1730 (1996).
Ebers, G. C., Yee, I. M., Sadovnick, A. D. & Duquette, P. Conjugal multiple sclerosis: population-based prevalence and recurrence risks in offspring. Ann. Neurol. 48, 927–931 (2000).
Brassat, D. et al. Familial factors influence disability in MS multiplex families. Neurology 52, 1632–1636 (1999).
Barcellos, L. F. et al. Genetic basis for clinical expression in multiple sclerosis. Brain 125, 150–158 (2002).
Hensiek, A. E. et al. Familial effects on the clinical course of multiple sclerosis. Neurology 68, 376–383 (2007).
DeLuca, G. C. et al. An extremes of outcome strategy provides evidence that multiple sclerosis severity is determined by alleles at the HLA-DRB1 locus. Proc. Natl Acad. Sci. USA 104, 20896–20901 (2007).
Willer, C. J., Dyment, D. A., Risch, N. J., Sadovnick, A. D. & Ebers, G. C. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc. Natl Acad. Sci. USA 100, 12877–12882 (2003).
Hansen, T. et al. Concordance for multiple sclerosis in Danish twins: an update of a nationwide study. Mult. Scler. 11, 504–510 (2005).
Hawkes, C. H. & Macgregor, A. J. Twin studies and the heritability of MS: a conclusion. Mult. Scler. 15, 661–667 (2009).
McElroy, J. P. & Oksenberg, J. R. Multiple sclerosis genetics. Curr. Top. Microbiol. Immunol. 318, 45–72 (2008).
Booth, D. R. et al. Gene expression and genotyping studies implicate the interleukin 7 receptor in the pathogenesis of primary progressive multiple sclerosis. J. Mol. Med. 83, 822–830 (2005).
Lundmark, F. et al. Variation in interleukin 7 receptor α chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 39, 1108–1113 (2007).
Gregory, S. G. et al. Interleukin 7 receptor α chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 39, 1083–1091 (2007).
Sawcer, S. et al. A high-density screen for linkage in multiple sclerosis. Am. J. Hum. Genet. 77, 454–467 (2005).
Burton, P. R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
International Multiple Sclerosis Genetics Consortium. Risk alleles for multiple sclerosis identified by a genome-wide study. N. Engl. J. Med. 357, 851–862 (2007).
Comabella, M. et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One 3, e3490 (2008).
Baranzini, S. E. et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 18, 767–778 (2009).
Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene). Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 41, 824–828 (2009).
Jakkula, E. et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am. J. Hum. Genet. 86, 285–291 (2010).
Sanna, S. et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat. Genet. 42, 495–497 (2010).
International Multiple Sclerosis Genetics Consortium (IMSGC). Refining genetic associations in multiple sclerosis. Lancet Neurol. 7, 567–569 (2008).
Ban, M. et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur. J. Hum. Genet. 17, 1309–1313 (2009).
D'Netto, M. J. et al. Risk alleles for multiple sclerosis in multiplex families. Neurology 72, 1984–1988 (2009).
Hoppenbrouwers, I. A. et al. Replication of CD58 and CLEC16A as genome-wide significant risk genes for multiple sclerosis. J. Hum. Genet. 54, 676–680 (2009).
International Multiple Sclerosis Genetics Consortium (IMSGC). The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 10, 11–14 (2009).
International Multiple Sclerosis Genetics Consortium (IMSGC). Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum. Mol. Genet. 19, 953–962 (2009).
De Jager, P. L. et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41, 776–782 (2009).
De Jager, P. L. et al. The role of the CD58 locus in multiple sclerosis. Proc. Natl Acad. Sci USA 106, 5264–5269 (2009).
Maier, L. M. et al. Soluble IL-2RA levels in multiple sclerosis subjects and the effect of soluble IL-2RA on immune responses. J. Immunol. 182, 1541–1547 (2009).
Baranzini, S. E. et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 18, 2078–2090 (2009).
Baranzini, S. E. The genetics of autoimmune diseases: a networked perspective. Curr. Opin. Immunol. 21, 596–605 (2009).
Hafler, J. P. et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 10, 5–10 (2009).
Sirota, M., Schaub, M. A., Batzoglou, S., Robinson, W. H. & Butte, A. J. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 5, e1000792 (2009).
Maier, L. M. & Hafler, D. A. The developing mosaic of autoimmune disease risk. Nat. Genet. 40, 131–132 (2008).
Dendrou, C. A. et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet. 41, 1011–1015 (2009).
Maier, L. M. et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 5, e1000322 (2009).
Goh, K. I. et al. The human disease network. Proc. Natl Acad. Sci. USA 104, 8685–8690 (2007).
Oti, M. & Brunner, H. G. The modular nature of genetic diseases. Clin. Genet. 71, 1–11 (2007).
Oti, M., Huynen, M. A. & Brunner, H. G. The biological coherence of human phenome databases. Am. J. Hum. Genet. 85, 801–808 (2009).
Loscalzo, J., Kohane, I. & Barabasi, A. L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol. Syst. Biol. 3, 124 (2007).
Marshall, C. R. et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008).
Wain, L. V., Armour, J. A. & Tobin, M. D. Genomic copy number variation, human health, and disease. Lancet 374, 340–350 (2009).
Zhang, F., Gu, W., Hurles, M. E. & Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481 (2009).
Schaschl, H., Aitman, T. J. & Vyse, T. J. Copy number variation in the human genome and its implication in autoimmunity. Clin. Exp. Immunol. 156, 12–16 (2009).
International Multiple Sclerosis Genetics Consortium (IMSGC). Evidence for polygenic susceptibility to multiple sclerosis. The shape of things to come. Am. J. Hum. Genet. 86, 621–625 (2010).
Altshuler, D., Daly, M. J. & Lander, E. S. Genetic mapping in human disease. Science 322, 881–888 (2008).
Marchini, J., Donnelly, P. & Cardon, L. R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 37, 413–417 (2005).
Lesnick, T. G. et al. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 3, e98 (2007).
Torkamani, A., Topol, E. J. & Schork, N. J. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics 92, 265–272 (2008).
Goldstein, D. B. Common genetic variation and human traits. N. Engl. J. Med. 360, 1696–1698 (2009).
Corvol, J. C. et al. Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc. Natl Acad. Sci. USA 105, 11839–11844 (2008).
De Jager, P. L. et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 8, 1111–1119 (2009).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J. A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009).
Dickson, S. P., Wang, K., Krantz, I., Hakonarson, H. & Goldstein, D. B. Rare variants create synthetic genome-wide associations. PLoS Biol. 8, e1000294 (2010).
Baranzini, S. E. et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464, 1351–1356 (2010).
Lupski, J. R. et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 362, 1181–1191 (2010).
Roach, J. C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Fugger, L., Friese, M. A. & Bell, J. I. From genes to function: the next challenge to understanding multiple sclerosis. Nat. Rev. Immunol. 9, 408–417 (2009).
Ascherio, A. & Munger, K. L. Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann. Neurol. 61, 504–513 (2007).
von Essen, M. R. et al. Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat. Immunol. 11, 344–349 (2010).
Correale, J., Ysrraelit, M. C. & Gaitan, M. I. Immunomodulatory effects of vitamin D in multiple sclerosis. Brain 132, 1146–1160 (2009).
Smolders, J. et al. Vitamin D status is positively correlated with regulatory T cell function in patients with multiple sclerosis. PLoS One 4, e6635 (2009).
Ramagopalan, S. V. et al. Expression of the multiple sclerosis-associated MHC class II allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 5, e1000369 (2009).
Becklund, B. R., Severson, K. S., Vang, S. V. & DeLuca, H. F. UV radiation suppresses experimental autoimmune encephalomyelitis independent of vitamin D production. Proc. Natl Acad. Sci. USA 107, 6418–6423 (2010).
Ascherio, A. & Munger, K. L. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann. Neurol. 61, 288–299 (2007).
Lipton, H. L., Liang, Z., Hertzler, S. & Son, K. N. A specific viral cause of multiple sclerosis: one virus, one disease. Ann. Neurol. 61, 514–523 (2007).
Ascherio, A. et al. Epstein–Barr virus antibodies and risk of multiple sclerosis: a prospective study. JAMA 286, 3083–3088 (2001).
Cepok, S. et al. Identification of Epstein–Barr virus proteins as putative targets of the immune response in multiple sclerosis. J. Clin. Invest. 115, 1352–1360 (2005).
Serafini, B. et al. Dysregulated Epstein–Barr virus infection in the multiple sclerosis brain. J. Exp. Med. 204, 2899–2912 (2007).
Salvetti, M., Giovannoni, G. & Aloisi, F. Epstein–Barr virus and multiple sclerosis. Curr. Opin. Neurol. 22, 201–206 (2009).
Riise, T., Nortvedt, M. W. & Ascherio, A. Smoking is a risk factor for multiple sclerosis. Neurology 61, 1122–1124 (2003).
Hedstrom, A. K., Baarnhielm, M., Olsson, T. & Alfredsson, L. Tobacco smoking, but not Swedish snuff use, increases the risk of multiple sclerosis. Neurology 73, 696–701 (2009).
Oksenberg, J. R. et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 74, 160–167 (2004).
Barcellos, L. F. et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am. J. Hum. Genet. 72, 710–716 (2003).
Dyment, D. A. et al. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum. Mol. Genet. 14, 2019–2026 (2005).
Gregersen, J. W. et al. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature 443, 574–577 (2006).
Caillier, S. J. et al. Uncoupling the roles of HLA-DRB1 and HLA–DRB5 genes in multiple sclerosis. J. Immunol. 181, 5473–5480 (2008).
Brynedal, B. et al. HLA-A confers an HLA-DRB1 independent influence on the risk of multiple sclerosis. PLoS One 2, e664 (2007).
Yeo, T. W. et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann. Neurol. 61, 228–236 (2007).
Friese, M. A. et al. Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nat. Med. 14, 1227–1235 (2008).
Lorentzen, A. R. et al. Killer immunoglobulin-like receptor ligand HLA-Bw4 protects against multiple sclerosis. Ann. Neurol. 65, 658–666 (2009).
Rioux, J. D. et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc. Natl Acad. Sci. USA 106, 18680–18685 (2009).
Hindorff, L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367 (2009).
Manolio, T. A. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI's office of population genomics. Pharmacogenomics 10, 235–241 (2009).
Ioannidis, J. P., Thomas, G. & Daly, M. J. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10, 318–329 (2009).
Manolio, T. A. et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009).
A Catalog of Published Genome-Wide Association Studies genome.gov [online], (2010).
Sadovnick, A. D. et al. Age of onset in concordant twins and other relative pairs with multiple sclerosis. Am. J. Epidemiol. 170, 289–296 (2009).
Byun, E. et al. Genome-wide pharmacogenomic analysis of the response to interferon beta therapy in multiple sclerosis. Arch. Neurol. 65, 337–344 (2008).
Comabella, M. et al. Genome-wide scan of 500,000 single-nucleotide polymorphisms among responders and nonresponders to interferon beta therapy in multiple sclerosis. Arch. Neurol. 66, 972–978 (2009).
Okuda, D. T. et al. Genotype–phenotype correlations in multiple sclerosis: HLA genes influence disease severity inferred by 1HMR spectroscopy and MRI measures. Brain 132, 250–259 (2009).
Pappas, D. J. & Oksenberg, J. R. Multiple sclerosis pharmacogenomics: maximizing efficacy of therapy. Neurology 74 (Suppl. 1), S62–S69 (2010).
Wu, J. S. et al. HLA-DRB1 allele heterogeneity influences multiple sclerosis severity as well as risk in Western Australia. J. Neuroimmunol. 219, 109–113 (2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Oksenberg, J., Baranzini, S. Multiple sclerosis genetics—is the glass half full, or half empty?. Nat Rev Neurol 6, 429–437 (2010). https://doi.org/10.1038/nrneurol.2010.91
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrneurol.2010.91
This article is cited by
-
What Have Failed, Interrupted, and Withdrawn Antibody Therapies in Multiple Sclerosis Taught Us?
Neurotherapeutics (2022)
-
Effects of Lactobacillus casei Strain T2 (IBRC-M10783) on the Modulation of Th17/Treg and Evaluation of miR-155, miR-25, and IDO-1 Expression in a Cuprizone-Induced C57BL/6 Mouse Model of Demyelination
Inflammation (2021)
-
Specialized Pro-Resolving Lipid Mediators: Emerging Therapeutic Candidates for Multiple Sclerosis
Clinical Reviews in Allergy & Immunology (2021)
-
An investigation of genetic polymorphisms in heparan sulfate proteoglycan core proteins and key modification enzymes in an Australian Caucasian multiple sclerosis population
Human Genomics (2020)
-
Outcomes and Cost-Effectiveness of Autologous Hematopoietic Cell Transplant for Multiple Sclerosis
Current Treatment Options in Neurology (2019)