Key Points
-
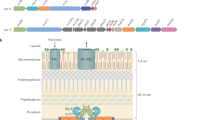
Type IV secretion systems (T4SSs) comprise 12 protein components, VirB1–VirB11 and VirD4. VirB2 forms a pilus and is co-purified with VirB5. VirB4, VirB11 and VirD4 are ATPases powering pilus assembly and substrate secretion. The inner membrane channel is probably composed of VirB6, VirB8 and VirB10. The composition of the outer membrane channel is still unknown, although it may include VirB9 and VirB7. The roles of VirB1 and VirB3 are unknown.
-
Over the past 10 years the structures of many T4SS components have been solved, including the recent elucidation of the structure of the core type IV secretion machine at 15 Å resolution using cryo-electron microscopy. The core complex from the conjugative plasmid pKM101 comprises 14 copies of the VirB7, VirB9 and VirB10 homologue proteins and spans the cell envelope. It is a cylindrical structure that is composed of two linked layers, the inner layer (I layer), which is anchored in the inner membrane, and the outer layer (O layer), which is inserted in the outer membrane.
-
The I layer is composed of the amino-terminal domains of the VirB9 and VirB10 homologues and has a 55 Å pore at its base. The O layer is composed of the VirB7 homologue and the carboxy-terminal domains of the VirB9 and VirB10 homologues, and consists of a main body and an outer cap that contains a small pore which allows communication with the extracellular milieu. This pore is not large enough for substrate secretion.
-
Various sub-assemblies are formed during type IV secretion; these are also described and their relevance in sub-assembly formation is discussed.
-
A general model for T4SS assembly is proposed. We suggest that VirB7, VirB9 and VirB10 assemble to form the core complex. VirB6 and VirB8 (which may form the inner membrane pore) and then the ATPase complex of VirB4 (possibly bound to VirB3), VirB11 and VirD4 dock onto the core complex to complete the translocation channel at the inner membrane. The VirB2 and VirB5 pilus subunits are then recruited to build either the distal portion of the secretion channel or the pilus.
-
The secretion pathway of T4SS substrates is discussed in the context of the structural knowledge of T4SS assembly. It was recently shown that the T-DNA substrate makes sequential contacts with VirD4, VirB11, VirB6, VirB8, VirB9 and VirB2, and a model is proposed that takes these data into account.
Abstract
Type IV secretion systems (T4SSs) are versatile secretion systems that are found in both Gram-negative and Gram-positive bacteria and secrete a wide range of substrates, from single proteins to protein–protein and protein–DNA complexes. They usually consist of 12 components that are organized into ATP-powered, double-membrane-spanning complexes. The structures of single soluble components or domains have been solved, but an understanding of how these structures come together has only recently begun to emerge. This Review focuses on the structural advances that have been made over the past 10 years and how the corresponding structural insights have helped to elucidate many of the details of the mechanism of type IV secretion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Grohmann, E., Muth, G. & Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301 (2003).
Lawley, T. D., Klimke, W. A., Gubbins, M. J. & Frost, L. S. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15 (2003).
Trieu-Cuot, P., Gerbaud, G., Lambert, T. & Courvalin, P. In vivo transfer of genetic information between Gram-positive and Gram-negative bacteria. EMBO J. 4, 3583–3587 (1985).
Bundock, P., den Dulk-Ras, A., Beijersbergen, A. & Hooykaas, P. J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214 (1995).
Heinemann, J. A. & Sprague, G. F. Jr. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340, 205–209 (1989).
Waters, V. L. Conjugation between bacterial and mammalian cells. Nature Genet. 29, 375–376 (2001).
Smeets, L. C. & Kusters, J. G. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 10, 159–162 (2002).
Hamilton, H. L. & Dillard, J. P. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59, 376–385 (2006).
Corbel, M. J. Brucellosis: an overview. Emerg. Infect. Dis. 3, 213–221 (1997).
Backert, S. & Meyer, T. F. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9, 207–217 (2006).
Ninio, S. & Roy, C. R. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 15, 372–380 (2007).
Lessl, M., Balzer, D., Pansegrau, W. & Lanka, E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J. Biol. Chem. 267, 20471–20480 (1992).
Lessl, M. & Lanka, E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77, 321–324 (1994).
Christie, P. J., Atmakuri, K., Krishnamoorthy, V., Jakubowski, S. & Cascales, E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 (2005).
Burns, D. L. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6, 29–34 (2003).
Seubert, A., Hiestand, R., de la Cruz, F. & Dehio, C. A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49, 1253–1266 (2003).
O'Callaghan, D. et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33, 1210–1220 (1999).
Gomis-Ruth, F. X. et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 (2001). This study reports the crystal structure of a VirD4 family protein, providing key insights into the molecular mechanisms of CP proteins.
Moncalian, G. et al. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274, 36117–36124 (1999).
Schroder, G. & Lanka, E. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185, 4371–4381 (2003).
Schroder, G. et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184, 2767–2779 (2002).
Tato, I., Zunzunegui, S., de la Cruz, F. & Cabezon, E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl Acad. Sci. USA 102, 8156–8161 (2005).
Tato, I. et al. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282, 25569–25576 (2007).
Hormaeche, I. et al. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 277, 46456–46462 (2002).
Gomis-Ruth, F. X., Moncalian, G., de la Cruz, F. & Coll, M. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277, 7556–7566 (2002).
Cao, T. B. & Saier, M. H. Jr. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147, 3201–3214 (2001).
Filloux, A. The type VI secretion system: a tubular story. EMBO J. 28, 309–310 (2009).
Planet, P. J., Kachlany, S. C., DeSalle, R. & Figurski, D. H. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl Acad. Sci. USA 98, 2503–2508 (2001).
Krause, S. et al. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl Acad. Sci. USA 97, 3067–3072 (2000).
Rashkova, S., Spudich, G. M. & Christie, P. J. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179, 583–591 (1997).
Krause, S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182, 2761–2770 (2000).
Rivas, S., Bolland, S., Cabezon, E., Goni, F. M. & de la Cruz, F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272, 25583–25590 (1997).
Savvides, S. N. et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22, 1969–1980 (2003).
Yeo, H. J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6, 1461–1472 (2000). This study reports the crystal structure of a VirB11 family protein and, together with reference 33, elucidates the nucleotide-dependent conformational changes that are required for its function.
Hare, S., Bayliss, R., Baron, C. & Waksman, G. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J. Mol. Biol. 360, 56–66 (2006).
Berger, B. R. & Christie, P. J. The Agrobacterium tumefaciens virB4 gene product is an essential virulence protein requiring an intact nucleoside triphosphate-binding domain. J. Bacteriol. 175, 1723–1734 (1993).
Cook, D. M., Farizo, K. M. & Burns, D. L. Identification and characterization of PtlC, an essential component of the pertussis toxin secretion system. Infect. Immun. 67, 754–759 (1999).
Fullner, K. J., Stephens, K. M. & Nester, E. W. An essential virulence protein of Agrobacterium tumefaciens, VirB4, requires an intact mononucleotide binding domain to function in transfer of T-DNA. Mol. Gen. Genet. 245, 704–715 (1994).
Rabel, C., Grahn, A. M., Lurz, R. & Lanka, E. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 185, 1045–1058 (2003).
Arechaga, I. et al. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J. Bacteriol. 190, 5472–5479 (2008).
Shirasu, K., Koukolikova-Nicola, Z., Hohn, B. & Kado, C. I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol. Microbiol. 11, 581–588 (1994).
Draper, O., Middleton, R., Doucleff, M. & Zambryski, P. C. Topology of the VirB4 C terminus in the Agrobacterium tumefaciens VirB/D4 type IV secretion system. J. Biol. Chem. 281, 37628–37635 (2006).
Dang, T. A. & Christie, P. J. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179, 453–462 (1997).
Yuan, Q. et al. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280, 26349–26359 (2005).
Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl Acad. Sci. USA 99, 11493–11500 (2002).
Terradot, L. et al. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol. Cell. Proteomics 3, 809–819 (2004).
Rain, J. C. et al. The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 (2001).
Malek, J. A. et al. Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica. Nucleic Acids Res. 32, 1059–1064 (2004).
Atmakuri, K., Cascales, E. & Christie, P. J. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54, 1199–1211 (2004).
Llosa, M., Zunzunegui, S. & de la Cruz, F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl Acad. Sci. USA 100, 10465–10470 (2003).
Gilmour, M. W., Lawley, T. D., Rooker, M. M., Newnham, P. J. & Taylor, D. E. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42, 705–715 (2001).
Jakubowski, S. J. et al. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71, 779–794 (2009).
Fronzes, R. et al. Structure of a type IV secretion system core complex. Science 323, 266–268 (2009). This study reports the first structure of a large T4SS assembly that is formed by the VirB7, VirB9 and VirB10 homologues that are encoded by the pKM101 conjugation plasmid.
Das, A. & Xie, Y. H. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27, 405–414 (1998).
Terradot, L. et al. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc. Natl Acad. Sci. USA 102, 4596–4601 (2005). This report describes the first structures of the periplasmic domains of two key T4SS components, VirB8 and VirB10.
Jakubowski, S. J., Cascales, E., Krishnamoorthy, V. & Christie, P. J. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J. Bacteriol. 187, 3486–3495 (2005).
Bayliss, R. et al. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl Acad. Sci. USA 104, 1673–1678 (2007). This article describes the first structure of a binary complex between the soluble, periplasmic parts of VirB9 and VirB7.
Cascales, E. & Christie, P. J. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl Acad. Sci. USA 101, 17228–17233 (2004).
Song, L. et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996).
Olson, R., Nariya, H., Yokota, K., Kamio, Y. & Gouaux, E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nature Struct. Biol. 6, 134–140 (1999).
Cascales, E. & Christie, P. J. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173 (2004). This study describes the interaction of a T4S substrate with the secretion machinery components.
Buhrdorf, R., Forster, C., Haas, R. & Fischer, W. Topological analysis of a putative virB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int. J. Med. Microbiol. 293, 213–217 (2003).
Kumar, R. B., Xie, Y. H. & Das, A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36, 608–617 (2000).
Judd, P. K., Kumar, R. B. & Das, A. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55, 115–124 (2005).
Baron, C. VirB8: a conserved type IV secretion system assembly factor and drug target. Biochem. Cell Biol. 84, 890–899 (2006).
Bailey, S., Ward, D., Middleton, R., Grossmann, J. G. & Zambryski, P. C. Agrobacterium tumefaciens VirB8 structure reveals potential protein-protein interaction sites. Proc. Natl Acad. Sci. USA 103, 2582–2587 (2006).
Karnholz, A. et al. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188, 882–893 (2006).
Jakubowski, S. J., Krishnamoorthy, V., Cascales, E. & Christie, P. J. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341, 961–977 (2004).
Kumar, R. B. & Das, A. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J. Bacteriol. 183, 3636–3641 (2001).
Krall, L. et al. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA 99, 11405–11410 (2002).
Jakubowski, S. J., Krishnamoorthy, V. & Christie, P. J. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185, 2867–2878 (2003).
Das, A. & Xie, Y. H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182, 758–763 (2000).
Rambow-Larsen, A. A. & Weiss, A. A. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 184, 2863–2869 (2002).
Hoppner, C., Carle, A., Sivanesan, D., Hoeppner, S. & Baron, C. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151, 3469–3482 (2005).
Bayer, M. et al. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 177, 4279–4288 (1995).
Koraimann, G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell. Mol. Life Sci. 60, 2371–2388 (2003).
Lai, E. M., Chesnokova, O., Banta, L. M. & Kado, C. I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182, 3705–3716 (2000).
Fullner, K. J., Lara, J. C. & Nester, E. W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273, 1107–1109 (1996).
Berger, B. R. & Christie, P. J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176, 3646–3660 (1994).
Bayer, M. et al. Functional and mutational analysis of P19, a DNA transfer protein with muramidase activity. J. Bacteriol. 183, 3176–3183 (2001).
Baron, C., Llosa, M., Zhou, S. & Zambryski, P. C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1. J. Bacteriol. 179, 1203–1210 (1997).
Zupan, J., Hackworth, C. A., Aguilar, J., Ward, D. & Zambryski, P. VirB1* promotes T-pilus formation in the vir-type IV secretion system of Agrobacterium tumefaciens. J. Bacteriol. 189, 6551–6563 (2007).
Beijersbergen, A., Smith, S. J. & Hooykaas, P. J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32, 212–218 (1994).
Grahn, A. M., Haase, J., Bamford, D. H. & Lanka, E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182, 1564–1574 (2000).
Batchelor, R. A., Pearson, B. M., Friis, L. M., Guerry, P. & Wells, J. M. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150, 3507–3517 (2004).
Christie, P. J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179, 3085–3094 (1997).
Haase, J., Lurz, R., Grahn, A. M., Bamford, D. H. & Lanka, E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 177, 4779–4791 (1995).
Babic, A., Lindner, A. B., Vulic, M., Stewart, E. J. & Radman, M. Direct visualization of horizontal gene transfer. Science 319, 1533–1536 (2008).
Shu, A. C. et al. Evidence of DNA transfer through F-pilus channels during Escherichia coli conjugation. Langmuir 24, 6796–6802 (2008).
Kwok, T. et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449, 862–866 (2007).
Eisenbrandt, R., Kalkum, M., Lurz, R. & Lanka, E. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol. 182, 6751–6761 (2000).
Sagulenko, E., Sagulenko, V., Chen, J. & Christie, P. J. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183, 5813–5825 (2001).
Haase, J. & Lanka, E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J. Bacteriol. 179, 5728–5735 (1997).
Majdalani, N. & Ippen-Ihler, K. Membrane insertion of the F-pilin subunit is Sec independent but requires leader peptidase B and the proton motive force. J. Bacteriol. 178, 3742–3747 (1996).
Majdalani, N., Moore, D., Maneewannakul, S. & Ippen-Ihler, K. Role of the propilin leader peptide in the maturation of F pilin. J. Bacteriol. 178, 3748–3754 (1996).
Moore, D. et al. The Escherichia coli K-12 F plasmid gene traX is required for acetylation of F pilin. J. Bacteriol. 175, 1375–1383 (1993).
Eisenbrandt, R. et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274, 22548–22555 (1999). A remarkable work describing the complex maturation process that T4SS pilins undergo during pilus biogenesis.
Kalkum, M., Eisenbrandt, R., Lurz, R. & Lanka, E. Tying rings for sex. Trends Microbiol. 10, 382–387 (2002).
Lai, E. M. & Kado, C. I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180, 2711–2717 (1998).
Schmidt-Eisenlohr, H. et al. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181, 7485–7492 (1999).
Schmidt-Eisenlohr, H., Domke, N. & Baron, C. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J. Bacteriol. 181, 5563–5571 (1999).
Yeo, H. J., Yuan, Q., Beck, M. R., Baron, C. & Waksman, G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl Acad. Sci. USA 100, 15947–15952 (2003). The crystal structure of a VirB5 protein suggests that it functions as an adhesin.
Aly, K. A. & Baron, C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology 153, 3766–3775 (2007).
Backert, S., Fronzes, R. & Waksman, G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 16, 409–413 (2008).
Bradley, D. E. Morphological and serological relationships of conjugative pili. Plasmid 4, 155–169 (1980).
Andrzejewska, J. et al. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J. Bacteriol. 188, 5865–5877 (2006).
Rohde, M., Puls, J., Buhrdorf, R., Fischer, W. & Haas, R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49, 219–234 (2003).
Tanaka, J., Suzuki, T., Mimuro, H. & Sasakawa, C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell. Microbiol. 5, 395–404 (2003).
Wang, Y. A., Yu, X., Silverman, P. M., Harris, R. L. & Egelman, E. H. The structure of F-pili. J. Mol. Biol. 385, 22–29 (2009).
Fernandez, D. et al. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178, 3156–3167 (1996).
Judd, P. K., Kumar, R. B. & Das, A. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl Acad. Sci. USA 102, 11498–11503 (2005).
Atmakuri, K., Ding, Z. & Christie, P. J. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49, 1699–1713 (2003).
Clarke, M., Maddera, L., Harris, R. L. & Silverman, P. M. F-pili dynamics by live-cell imaging. Proc. Natl Acad. Sci. USA 105, 17978–17981 (2008).
Filloux, A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694, 163–179 (2004).
Koronakis, V., Hughes, C. & Koronakis, E. Energetically distinct early and late stages of HlyB/HlyD-dependent secretion across both Escherichia coli membranes. EMBO J. 10, 3263–3272 (1991).
Omori, K. & Idei, A. Gram-negative bacterial ATP-binding cassette protein exporter family and diverse secretory proteins. J. Biosci. Bioeng. 95, 1–12 (2003).
Gold, V. A., Duong, F. & Collinson, I. Structure and function of the bacterial Sec translocon. Mol. Membr. Biol. 24, 387–394 (2007).
Voulhoux, R. et al. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20, 6735–6741 (2001).
Peabody, C. R. et al. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072 (2003).
Craig, L. & Li, J. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18, 267–277 (2008).
Cornelis, G. R. & Van Gijsegem, F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774 (2000).
Galan, J. E. & Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 (2006).
Henderson, I. R., Navarro-Garcia, F., Desvaux, M., Fernandez, R. C. & Ala'Aldeen, D. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68, 692–744 (2004).
Henderson, I. R. & Nataro, J. P. Virulence functions of autotransporter proteins. Infect. Immun. 69, 1231–1243 (2001).
Mazar, J. & Cotter, P. A. New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 15, 508–515 (2007).
Cascales, E. The type VI secretion toolkit. EMBO Rep. 9, 735–741 (2008).
Pukatzki, S., McAuley, S. B. & Miyata, S. T. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 (2009).
Cascales, E. & Christie, P. J. The versatile bacterial type IV secretion systems. Nature Rev. Microbiol. 1, 137–149 (2003).
Dehio, C. Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell. Microbiol. 10, 1591–1598 (2008).
Sagulenko, V., Sagulenko, E., Jakubowski, S., Spudich, E. & Christie, P. J. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183, 3642–3651 (2001).
Jones, A. L., Shirasu, K. & Kado, C. I. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176, 5255–5261 (1994).
Strauch, E. et al. A cryptic plasmid of Yersinia enterocolitica encodes a conjugative transfer system related to the regions of CloDF13 Mob and IncX Pil. Microbiology 149, 2829–2845 (2003).
Shamaei-Tousi, A., Cahill, R. & Frankel, G. Interaction between protein subunits of the type IV secretion system of Bartonella henselae. J. Bacteriol. 186, 4796–4801 (2004).
Anderson, L. B., Hertzel, A. V. & Das, A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl Acad. Sci. USA 93, 8889–8894 (1996).
Baron, C., Thorstenson, Y. R. & Zambryski, P. C. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179, 1211–1218 (1997).
Das, A., Anderson, L. B. & Xie, Y. H. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 179, 3404–3409 (1997).
Farizo, K. M., Cafarella, T. G. & Burns, D. L. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271, 31643–31649 (1996).
Harris, R. L., Hombs, V. & Silverman, P. M. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42, 757–766 (2001).
Spudich, G. M., Fernandez, D., Zhou, X. R. & Christie, P. J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl Acad. Sci. USA 93, 7512–7517 (1996).
Beaupre, C. E., Bohne, J., Dale, E. M. & Binns, A. N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179, 78–89 (1997).
Acknowledgements
This work was funded by Wellcome Trust grant 082227 to G.W. and National Institutes of Health grant GM48746 to P.J.C.
Author information
Authors and Affiliations
Corresponding authors
Related links
Related links
DATABASES
Entrez Genome Project
Salmonella enterica subsp. enterica serovar Typhimurium
FURTHER INFORMATION
Glossary
- General secretory pathway
-
The pathway in which substrates are targeted and secreted through the Sec machinery.
- Walker A and B motif
-
A Walker A motif is an amino acid motif (GXXXGKT, in which X denotes any amino acid residue) that is involved in the nucleotide binding that occurs in many ATP-requiring enzymes. A Walker B motif is also a conserved sequence (usually XXXXD, in which X denotes any hydrophobic amino acid residue) that is used in conjunction with the Walker A motif to hydrolyse ATP.
- T-DNA
-
One of the substrates of the T4SS encoded by the Vir system of Agrobacterium tumefaciens. T-DNA is encoded by the A. tumefaciens Ti plasmid and is essential for A. tumefaciens pathogenesis.
- Type IV pilus
-
An elongated, flexible appendage that extends from the surface of Gram-negative bacterial cells and is used for adhesion and for cell motility (twitching motility).
- F-pilus
-
The pilus formed by the T4SS that is encoded by the F plasmid.
- Relaxase
-
A protein that targets the origin of the transfer sequence on plasmid DNA. Along with other proteins, it targets the DNA to the T4SS.
Rights and permissions
About this article
Cite this article
Fronzes, R., Christie, P. & Waksman, G. The structural biology of type IV secretion systems. Nat Rev Microbiol 7, 703–714 (2009). https://doi.org/10.1038/nrmicro2218
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2218
This article is cited by
-
Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer
Nature Communications (2023)
-
Current advances in microbial-based cancer therapies
Medical Oncology (2023)
-
Proteomic analysis of Rickettsia akari proposes a 44 kDa-OMP as a potential biomarker for Rickettsialpox diagnosis
BMC Microbiology (2020)
-
Parallel reductive genome evolution in Desulfovibrio ectosymbionts independently acquired by Trichonympha protists in the termite gut
The ISME Journal (2020)
-
Comprehensive genomic analysis of an indigenous Pseudomonas pseudoalcaligenes degrading phenolic compounds
Scientific Reports (2019)