Key Points
-
Bacterial type IV secretion (T4S) systems are ancestrally related to conjugation systems. Present-day T4S systems can be subclassified into three groups: conjugation systems mediating the transfer of DNA and protein substrates through formation of stable mating junctions with recipient cells; DNA-uptake and -release systems that exchange DNA with the extracellular milieu; and effector translocator systems that deliver DNA and protein effector molecules to eukaryotic cells during the course of infection.
-
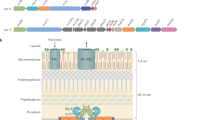
The conjugation systems of Gram-negative bacteria are composed of the coupling protein homomultimer and two substructures encoded by the mating-pair-formation (Mpf) proteins, a protein complex spanning the cell envelope and the conjugative pilus (T-pilus).
-
At the present time, investigators have identified the effector molecules of T4S systems for four pathogens: Agrobacterium tumefaciens, the causative agent of crown gall disease in plants; Helicobacter pylori, the causative agent of gastritis, peptic ulcers and gastric carcinomas in humans; Bordetella pertussis, the causative agent of whooping cough in humans; and Legionella pneumophila, the causative agent of Legionnaire's disease in humans.
-
T4S system effector translocation alters a wide range of cellular processes to aid the infection process. For example, Agrobacterium tumefaciens delivers oncogenic T-DNA and protein effectors (VirE2, VirE3 and VirF) that interact with plant cellular factors to ensure delivery of the T-DNA to the nucleus and its integration into the plant genome.
-
Helicobacter pylori uses the Cag T4S system to deliver CagA to mammalian cells where it is tyrosine phosphorylated by c-Src kinase. A complex network of interactions between CagAP-Tyr and cellular factors (for example, c-Src, SHP-2, c-MET) and between non-phosphorylated CagA (Grb2, JAM, Z0-1) alters signalling pathways, ultimately triggering cellular proliferation and differentiation and the onset of cancer.
-
The Bordetella pertussis Ptl system delivers its effector, the multisubunit pertussis toxin, to the extracellular milieu where, on contact with the mammalian cell membrane, the A subunit is internalized and exerts its effects by uncoupling G proteins from their receptors.
-
In striking contrast to the other systems described in the review, the intracellular pathogen L. pneumophila uses the Dot/Icm T4S system to inject effectors (DotA, LidA, RalF) into the phagosome to control biogenesis of the replicative vacuoule and to modulate the activities of host factors involved in vesicle traffic.
Abstract
Bacteria use type IV secretion systems for two fundamental objectives related to pathogenesis — genetic exchange and the delivery of effector molecules to eukaryotic target cells. Whereas gene acquisition is an important adaptive mechanism that enables pathogens to cope with a changing environment during invasion of the host, interactions between effector and host molecules can suppress defence mechanisms, facilitate intracellular growth and even induce the synthesis of nutrients that are beneficial to bacterial colonization. Rapid progress has been made towards defining the structures and functions of type IV secretion machines, identifying the effector molecules, and elucidating the mechanisms by which the translocated effectors subvert eukaryotic cellular processes during infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cavelli-Sforza, L., Lederberg, J. & Lederberg. E. An infective factor controlling sex compatibility in B. coli. J. Gen. Microbiol. 8, 89–103 (1953).
Lawley, T. D., Klimke, W. A., Gubbins, M. J. & Frost, L. S. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224, 1–15 (2003).
Winans, S. C., Burns, D. L. & Christie, P. J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4, 64–68 (1996).
Censini, S. et al. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl Acad. Sci. USA 93, 14648–14653 (1996).
Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279, 873–876 (1998). Reports the discovery of a type IV secretion system that is evolutionarily related to the IncI plasmids and functions dually in conjugative transfer and effector translocation.
Bacon, D. J. et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect. Immun. 68, 4384–4390 (2000).
Hofreuter, D., Odenbreit, S. & Haas, R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41, 379–391 (2001).
Dillard, J. P. & Seifert, H. S. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41, 263–277 (2001). References 6–8 report the discoveries of new type IV translocation systems, herein termed the 'DNA uptake and release' subfamily.
Bundock, P., den Dulk Ras, A., Beijersbergen, A. & Hooykaas, P. J. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214 (1995).
Bates, S., Cashmore, A. M. & Wilkins, B. M. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae. J. Bacteriol. 180, 6538–6543 (1998).
Zhu, J. et al. The bases of crown gall tumorigenesis. J. Bacteriol. 182, 3885–3895 (2000).
Waters, V. L. Conjugation between bacterial and mammalian cells. Nature Genet. 29, 375–376 (2001).
Grohmann, E., Muth, G. & Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301 (2003).
de la Cruz, I. & Davies, J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8, 128–133 (2000).
Ghigo, J. M. Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445 (2001).
Schlievert, P. M. et al. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66, 218–223 (1998).
Bacon, D. J. et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81–176. Infect. Immun. 70, 6242–6250 (2002).
Hofreuter, D., Odenbreit, S., Puls, J., Schwan, D. & Haas, R. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 151, 487–491 (2000).
Hamilton, H. L., Schwartz, K. J. & Dillard, J. P. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 183, 4718–4726 (2001).
Chen, I. & Dubnau, D. DNA transport during transformation. Front. Biosci. 8, S544–S556 (2003).
Blocker, A., Komoriya, K. & Aizawa, S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl Acad. Sci. USA 100, 3027–3030 (2003).
Christie, P. J. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179, 3085–3094 (1997).
Hilbi, H., Segal, G. & Shuman, H. A. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42, 603–617 (2001).
Schmiederer, M., Arcenas, R., Widen, R., Valkov, N. & Anderson, B. Intracellular induction of the Bartonella henselae virB operon by human endothelial cells. Infect. Immun. 69, 6495–6502 (2001).
Boschiroli, M. L. et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl Acad. Sci. USA 99, 1544–1549 (2002).
Seubert, A., Hiestand, R., de la Cruz, F. & Dehio, C. A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49, 1253–1266 (2003).
Rouot, B. et al. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71, 1075–1082 (2003).
Baron, C., O'Callaghan, D. & Lanka, E. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43, 1359–1365 (2002).
Llosa, M., Gomis-Ruth, F. X., Coll, M. & de la Cruz, F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45, 1–8 (2002).
Gomis-Ruth, F. X. & Coll, M. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33, 839–843 (2001).
Gomis-Ruth, F. X. et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 (2001). Reports the first structure of a coupling protein, a hexameric F1-ATPase-like multimer that is highly indicative of a translocase function.
Hormaeche, I. et al. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 277, 46456–46462 (2002).
Errington, J., Bath, J. & Wu, L. J. DNA transport in bacteria. Nature Rev. Mol. Cell Biol. 2, 538–545 (2001).
Schroder, G. et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184, 2767–2779 (2002).
Gilmour, M. W., Gunton, J. E., Lawley, T. D. & Taylor, D. E. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49, 105–116 (2003).
Llosa, M., Zunzunegui, S. & de la Cruz, F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl Acad. Sci. USA 100, 10465–10470 ( 2003).
Kumar, R. B. & Das, A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43, 1523–1532 (2002).
Ding, Z. et al. A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J. Bacteriol. 184, 5572–5582 (2002).
Atmakuri, K., Ding, Z. & Christie, P. J. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at the cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49, 1699–1713 (2003). Reports the first direct evidence for recruitment of a protein effector to a coupling protein, supporting a proposal that the coupling proteins function as general recruitment factors for type IV secretion substrates.
Vergunst, A. C. et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 (2000). This elegant study supplied the first incontrovertible evidence for protein trafficking by the A. tumefaciens VirB/D4 type IV secretion system.
Simone, M., McCullen, C. A., Stahl, L. E. & Binns, A. N. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 41, 1283–1293 (2001).
Schrammeijer, B., den Dulk-Ras A, Vergunst, A. C., Jurado Jacome, E. & Hooykaas, P. J. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucl. Acids Res. 31, 860–868 (2003).
Selbach, M., Moese, S., Meyer, T. F. & Backert, S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect. Immun. 70, 665–667 (2002).
Conover, G. M., Derre, I., Vogel, J. P. & Isberg, R. R. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48, 305–321 (2003). Describes the discovery of a novel effector, LidA, that is required for elaboration of the L. pneumophila Dot/Icm complex in the bacterium and, on translocation, functions in vesicle recruitment during biogenesis of the replication vacuole.
Burns, D. L. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6, 29–34 (2003).
O'Callaghan, D. et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33, 1210–1220 (1999).
Lai, E. M. & Kado, C. I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8, 361–369 (2000).
Eisenbrandt, R. et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274, 22548–22555 (1999). Provides evidence that the pilin subunits of the A. tumefaciens VirB/D4 and the plasmid RP4 Tra systems undergo head-to-tail cyclization during maturation, a completely new reaction in bacteria.
Schmidt-Eisenlohr, H. et al. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181, 7485–7492 (1999).
Sagulenko, V., Sagulenko, E., Jakubowski, S., Spudich, E. & Christie, P. J. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183, 3642–3651 (2001).
Baron, C., Llosa, M., Zhou, S. & Zambryski, P. C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 179, 1203–1210 (1997).
Berger, B. R. & Christie, P. J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176, 3646–3660 (1994).
Kumar, R. B., Xie, Y. H. & Das, A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36, 608–617 (2000).
Das, A., Anderson, L. B. & Xie, Y. H. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 179, 3404–3409 (1997).
Beauprè, C. E., Bohne, J., Dale, E. M. & Binns, A. N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179, 78–89 (1997).
Das, A. & Xie, Y. -H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182, 758–763 (2000).
Ward, D. V., Draper, O., Zupan, J. R. & Zambryski, P. C. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl Acad. Sci. USA 99, 11493–11500 (2002). Reports a comprehensive linkage map of the VirB proteins by yeast dihybrid screens.
Krall, L. et al. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA 99, 11405–11410 (2002).
Jakubowski, S. J., Krishnamoorthy, V. & Christie, P. J. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185, 2867–2878 (2003).
Rambow-Larsen, A. A. & Weiss, A. A. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 184, 2863–2869 (2002).
Farizo, K. M., Cafarella, T. G. & Burns, D. L. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI–PtlF complex. J. Biol. Chem. 271, 31643–31649 (1996).
Harris, R. L., Hombs, V. & Silverman, P. M. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42, 757–766 (2001).
Liu, Z. & Binns, A. N. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 185, 3259–3269 (2003).
Planet, P. J., Kachlany, S. C., DeSalle, R. & Figurski, D. H. Phylogeny of genes for secretion NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl Acad. Sci. USA 98, 2503–2508 (2001).
Yeo, H. -J., Savvides, S. N., Herr, A. B., Lanka, E. & Waksman, G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol. Cell 6, 1461–1472 (2000). Reports the first structure of a member of the VirB11 superfamily of traffic ATPases.
Savvides, S. N. et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22, 1969–1980 (2003).
Rabel, C., Grahn, A. M., Lurz, R. & Lanka, E. The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 185, 1045–1058 (2003).
Dang, T. A., Zhou, X. -R., Graf, B. & Christie, P. J. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol. Microbiol. 32, 1239–1251 (1999).
Sagulenko, Y., Sagulenko, V., Chen, J. & Christie, P. J. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183, 5813–5825 (2001).
Rohde, M., Puls, J., Buhrdorf, R., Fischer, W. & Haas, R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49, 219–234 (2003).
Tanaka, J., Suzuki, T., Mimuro, H. & Sasakawa, C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell. Microbiol. 5, 395–404 (2003). References 70 and 71 report that the Cag type IV secretion system elaborates new sheathed surface structures that contribute to H. pylori pathogenesis.
Schulein, R. & Dehio, C. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46, 1053–1067 (2002).
Watarai, M., Andrews, H. L. & Isberg, R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 39, 313–329 (2001).
Farizo, K. M., Fiddner, S., Cheung, A. M. & Burns, D. L. Membrane localization of the S1 subunit of pertussis toxin in Bordetella pertussis and implications for pertussis toxin secretion. Infect. Immun. 70, 1193–1201 (2002).
Grahn, A. M., Haase, J., Bamford, D. H. & Lanka, E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182, 1564–1574 (2000).
Pantoja, M., Chen, L., Chen, Y. & Nester, E. W. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 45, 1325–1335 (2002).
Galan, J. E. & Collmer, A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284, 1322–1328 (1999).
Ward, D. V. & Zambryski, P. C. The six functions of Agrobacterium VirE2. Proc. Natl Acad. Sci. USA 98, 385–386 (2001).
Deng, W. et al. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl Acad. Sci. USA 95, 7040–7045 (1998).
Gelvin, S. B. Agrobacterium-mediated plant transformation: the biology behind the 'gene-jockeying' tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003).
Tzfira, T. & Citovsky, V. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12, 121–129 (2002).
Zhu, Y. et al. Identification of Arabidopsis rat mutants. Plant Physiol. 132, 494–505 (2003).
Roberts, R. L. et al. Purine synthesis and increased Agrobacterium tumefaciens transformation of yeast and plants. Proc. Natl Acad. Sci. USA 100, 6634–6639 (2003).
Segal, E., Cha, J., Lo, J., Falkow, S. & Tompkins, L. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl Acad. Sci. USA 96, 14559–14564 (1999).
Hatakeyama, M. Helicobacter pylori CagA — a potential bacterial oncoprotein that functionally mimics the mammalian Gab family of adaptor proteins. Microbes Infect. 5, 143–150 (2003).
Asahi, M. et al. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 191, 593–602 (2000).
Stein, M., Rappuoli, R. & Covacci, A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl Acad. Sci. USA 97, 1263–1268 (2000).
Odenbreit, S. et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287, 1497–1500 (2000).
Stein, M. et al. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43, 971–980 (2002).
Backert, S. et al. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2, 155–164 (2000). References 84, 86, 87, 88 and 90 report the first evidence for CagA translocation and subsequent phosphorylation in human cells.
Selbach, M., Moese, S., Hauck, C. R., Meyer, T. F. & Backert, S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277, 6775–6778 (2002).
Higashi, H. et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295, 683–686 (2002).
Churin, Y. et al. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161, 249–255 (2003).
Mimuro, H. et al. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 10, 745–755 (2002).
Amieva, M. R. et al. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300, 1430–1434 (2003). Shows that non-phosphorylated CagA induces formation of an aberrant apical-junction protein complex, resulting in loss of cell polarity, proliferation and differentiation.
Fischer, W. et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42, 1337–1348 (2001).
Stein, P. E. et al. The crystal structure of pertussis toxin. Structure 2, 45–57 (1994).
Albert, P. R. & Robillard, L. G-protein specificity: traffic direction required. Cell Signal. 14, 407–418 (2002).
Vogel, J. P. & Isberg, R. R. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2, 30–34 (1999).
Swanson, M. S. & Sturgill-Koszycki, S. Exploitation of macrophages as a replication niche by Legionella pneumophila. Trends Microbiol. 8, 47–49 (2000).
Segal, G., Purcell, M. & Schuman, H. A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl Acad. Sci. USA 95, 1669–1674 (1998).
Segal, G. & Shuman, H. A. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 30, 197–208 (1998).
Nagai, H. & Roy, C. R. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20, 5962–5970 (2001).
Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679–682 (2002).
Boschiroli, M. L. et al. Type IV secretion and Brucella virulence. Vet. Microbiol. 90, 341–348 (2002).
Novak, K. F., Dougherty, B. & Pelaez, M. Actinobacillus actinomycetemcomitans harbours type IV secretion system genes on a plasmid and in the chromosome. Microbiology 147, 3027–3035 (2001).
Ohashi, N., Zhi, N., Lin, Q. & Rikihisa, Y. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70, 2128–2138 (2002).
Masui, S., Sasaki, T. & Ishikawa, H. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J. Bacteriol. 182, 6529–6531 (2000).
Andersson, S. G. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140 (1998).
Bhattacharyya, A. et al. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl Acad. Sci. USA 99, 12403–12408 (2002).
Zusman, T., Yerushalmi, G. & Segal, G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect. Immun. 71, 3714–3723 (2003).
Burghdorf, R., Forster, C., Haas, R. & Fischer, W. Topological analysis of a putative VirB8 homologue essential for the cag type IV secretion system in Helicobacter pylori. Int. J. Med. Microbiol. 293, 213–217 (2003).
Cao, T. B. & Saier, M. H. Jr. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints, and evolutionary conclusions. Microbiology 147, 3201–3214 (2001).
Acknowledgements
We dedicate this review to Brian Wilkins in loving memory. We apologize for any omissions in citation of primary reports owing to space limitations. We thank members of the laboratory for helpful comments and critical appraisals of this manuscript. We also gratefully acknowledge the National Institutes of Health for supporting our studies of the Agrobacterium VirB/D4 T4S system.
Author information
Authors and Affiliations
Corresponding author
Glossary
- TYPE IV SECRETION (T4S) APPARATUS OR SYSTEM
-
A bacterial organelle that is ancestrally related to a conjugation machine that translocates DNA or protein substrates across the cell envelope, often for purposes associated with pathogenesis. Other bacterial translocation systems include the type II secretion (T2S) machines that deliver protein substrates across the outer membrane and the type III secretion (T3S) machines that translocate effectors in one step across the cell envelope through a structure that is ancestrally related to the bacterial flagellum. Like the T4S systems, the T3S systems elaborate syringe- or pilus-like surface organelles and deliver effector proteins to plant and mammalian cells during infection.
- CONJUGATION
-
A mechanism for transfer of a DNA substrate from a bacterial donor cell to a recipient cell by direct cell-to-cell contact.
- PILUS
-
A filamentous organelle that extends from the surface of the bacterial cell. Composed of pilin subunits, these structures mediate attachment to target cells or inert matter. They might also participate directly in the delivery of secretion substrates to target cells.
- COMPETENCE
-
The ability of a bacterial cell to import exogenous DNA and stably incorporate it into the bacterial genome.
- PERIPLASM
-
An aqueous compartment between the inner and outer membranes of Gram-negative bacteria.
- CHAPERONE
-
A protein or protein complex that participates in folding or unfolding of protein substrates. Secretion chaperones prevent their substrates from aggregating or interacting prematurely with other substrates or cellular factors. They might also mediate the delivery of substrates to a secretory apparatus.
- SECRETION SIGNAL
-
A motif that confers recognition of a protein that is destined for export by a cognate secretory apparatus.
- GENERAL SECRETORY PATHWAY
-
(GSP). The main pathway used for delivery of protein substrates into or across the bacterial inner membrane.
- INTERLEUKIN-8
-
(IL-8). A peptide that is produced by epithelial cells and is an indicator of infection. IL-8 secretion is induced by pathogens preceding clinical complications.
- TRANSGLYCOSYLASE
-
A protein, the enzymatic activity of which degrades peptidoglycan. These proteins participate in assembly of supramolecular transenvelope structures by 'punching' holes in the peptidoglycan.
- PEPTIDOGLYCAN
-
A shape-determining polymer that is present within the periplasm of Gram-negative bacteria.
- SECRETIN
-
A protein that forms oligomeric pores to allow the passage of macromolecular substrates across the outer membrane.
- KARYOPHERIN
-
These proteins have a central role in nuclear import processes, mediating substrate recognition and release at the nuclear-pore complex by GTP-hydrolysis-dependent reactions.
- CORTACTIN
-
Cortactin is an actin-binding protein, regulated by the membrane-associated c-Src kinase. Cortactin transduces signals from the cell surface to the cytoskeleton.
- ACTIN
-
A eukaryotic protein that polymerizes to form microfilaments. Microfilaments have a dual role, acting as a passive structural complex that maintains cell shape and anchors cytoskeletal proteins, and an active function for the transport of vesicles and organelles that can result in cell movement.
- APICAL JUNCTION
-
Apical junctions consist of protein complexes that join the actin cytoskeleton to the apical pole of epithelial cells. Adherent junctions have pivotal roles in cell organization by mediating cell adhesion and signalling.
- ADP RIBOSYLATION FACTOR
-
(ARF). The ADP-ribosylation factor family of small GTP-binding proteins is involved in the regulation of membrane traffic (vesicular transport) and in the control of the actin cytoskeleton.
Rights and permissions
About this article
Cite this article
Cascales, E., Christie, P. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1, 137–149 (2003). https://doi.org/10.1038/nrmicro753
Issue Date:
DOI: https://doi.org/10.1038/nrmicro753
This article is cited by
-
Structural and functional diversity of type IV secretion systems
Nature Reviews Microbiology (2024)
-
Systematic investigation of recipient cell genetic requirements reveals important surface receptors for conjugative transfer of IncI2 plasmids
Communications Biology (2023)
-
Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nature Communications (2022)
-
Genome analysis of Spiroplasma citri strains from different host plants and its leafhopper vectors
BMC Genomics (2021)
-
The complete genome of 2,6-dichlorobenzamide (BAM) degrader Aminobacter sp. MSH1 suggests a polyploid chromosome, phylogenetic reassignment, and functions of plasmids
Scientific Reports (2021)