Key Points
-
Stem cells are endowed with self-renewal and multi-lineage differentiation potential that enable them to generate mature functional tissues during development and to regenerate these tissues following injury or degenerative processes.
-
The regulation of stem cells is thought to be mediated by the convergence of extrinsic (microenvironmental) and intrinsic (intracellular) signals that currently are poorly understood.
-
The genetic regulation of embryonic and adult stem cells is studied using focused and global techniques for gene-expression analysis.
-
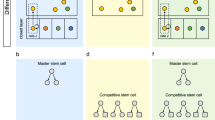
It has been shown that stem cells express many lineage-specific genes at low levels before lineage commitment, and specification to a single lineage is not only because of acquisition of lineage-specific gene-expression during differentiation, but is also associated with loss of promiscuous gene expression.
-
The many reported embryonic and adult stem-cell transcriptomes have provided a foundation for understanding the molecular mechanisms that regulate the stem-cell phenotype, but discordant results from these studies have prevented the identification of a conserved stem-cell molecular signature, if such an entity exists.
-
The ability to prospectively purify stem-cell populations to homogeneity, and to therefore eliminate the contaminating transcripts from committed cells, will advance our understanding of stem-cell gene expression, and combined with rapid high-throughput functional-genomics approaches, should reveal the molecular mechanisms that underlie the stem-cell phenotype.
-
As gene expression does not necessarily dictate which of the genes or gene pathways are functionally involved in self-renewal and pluri-/multi-potency, the next step to define the implications of an expressed gene pattern will be to design gene-targeting experiments to characterize the function of these genes and gene pathways.
Abstract
Stem cells share the defining characteristics of self-renewal, which maintains or expands the stem-cell pool, and multi-lineage differentiation, which generates and regenerates tissues. Stem-cell self-renewal and differentiation are influenced by the convergence of intrinsic cellular signals and extrinsic microenvironmental cues from the surrounding stem-cell niche, but the specific signals involved are poorly understood. Recently, several studies have sought to identify the genetic mechanisms that underlie the stem-cell phenotype. Such a molecular road map of stem-cell function should lead to an understanding of the true potential of stem cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jiang, Y. et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41–49 (2002).
Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Labosky, P. A., Barlow, D. P. & Hogan, B. L. Embryonic germ cell lines and their derivation from mouse primordial germ cells. Ciba. Found. Symp. 182, 157–168; discussion 168–178 (1994).
Pain, B. et al. Long-term in vitro culture and characterisation of avian embryonic stem cells with multiple morphogenetic potentialities. Development 122, 2339–2348 (1996).
Thomson, J. A. & Marshall, V. S. Primate embryonic stem cells. Curr. Top. Dev. Biol. 38, 133–165 (1998).
Ogawa, M. Differentiation and proliferation of hematopoietic stem cells. Blood 81, 2844–2853 (1993).
Ivanova, N. B. et al. A stem cell molecular signature. Science 298, 601–604 (2002).
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C. & Melton, D. A. 'Stemness': transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600 (2002). References 8 and 9 are the first analyses of global gene-expression profiling of multiple adult and embryonic stem-cell populations to identify conserved stem-cell genes.
Fortunel, N. O. et al. Comment on “'Stemness': transcriptional profiling of embryonic and adult stem cells” and “A stem cell molecular signature”. Science 302, 393; author reply 393 (2003). Similar to references 8 and 9, reports another global gene-expression analysis of multiple adult and embryonic stem-cell populations, and compares the stem-cell genes identified in references 8–10 pointing out the discordance of these studies.
Abeyta, M. J. et al. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum. Mol. Genet. 13, 601–608 (2004). This extensive work identifies genes expressed in multiple human ESC lines as well as comparing these genes to mouse ES and adult stem-cell profiles.
Sato, N. et al. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 260, 404–413 (2003).
Sperger, J. M. et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl Acad. Sci. USA 100, 13350–13355 (2003).
Brandenberger, R. et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nature Biotechnol. 22, 707–716 (2004).
Hu, M. et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 (1997).
Terskikh, A. V., Miyamoto, T., Chang, C., Diatchenko, L. & Weissman, I. L. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood 102, 94–101 (2003).
Akashi, K. et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101, 383–389 (2003).
Ye, M. et al. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity 19, 689–699 (2003).
Arney, K. L. & Fisher, A. G. Epigenetic aspects of differentiation. J. Cell Sci. 117, 4355–4363 (2004).
Tanaka, T. S. et al. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 12, 1921–1928 (2002).
Sharov, A. A. et al. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 1, E74 (2003).
Nichols, J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998).
Chambers, I. et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 (2003).
Mitsui, K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003).
Rao, M. Conserved and divergent paths that regulate self-renewal in mouse and human embryonic stem cells. Dev. Biol. 275, 269–286 (2004).
Niwa, H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 26, 137–148 (2001). An excellent review on OCT4 signalling in ESCs that describes the achievements and the difficulties of confirming gene function in stem cells.
Chambers, I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells 6, 386–391 (2004).
Richards, M., Tan, S. P., Tan, J. H., Chan, W. K. & Bongso, A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells 22, 51–64 (2004).
Bhattacharya, B. et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood 103, 2956–2964 (2004).
Brandenberger, R. et al. MPSS profiling of human embryonic stem cells. BMC Dev. Biol. 4, 10 (2004).
Ying, Q. L., Nichols, J., Chambers, I. & Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 (2003).
Xu, R. H. et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature Biotechnol. 20, 1261–1264 (2002).
Xu, R. H. et al. Basic FGF and supression of BMP signaling sustain undifferentiated proliferation of human ES cells. Nature Methods 2, 185–190 (2005).
Osawa, M., Hanada, K., Hamada, H. & Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996).
Ellisen, L. W. et al. TAN-1, the human homolog of the Drosophila Notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66, 649–661 (1991).
Begley, C. G. et al. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor δ-chain diversity region and results in a previously unreported fusion transcript. Proc. Natl Acad. Sci. USA 86, 2031–2035 (1989).
Finger, L. R. et al. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc. Natl Acad. Sci. USA 86, 5039–5043 (1989).
Royer-Pokora, B., Loos, U. & Ludwig, W. D. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene 6, 1887–1893 (1991).
Varnum-Finney, B. et al. Pluripotent, cytokine-dependent, hematopoietic stem cells are immortalized by constitutive Notch1 signaling. Nature Med. 6, 1278–1281 (2000).
Lecuyer, E. & Hoang, T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32, 11–24 (2004).
Yamada, Y. et al. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl Acad. Sci. USA 95, 3890–3895 (1998).
Varnum-Finney, B., Brashem-Stein, C. & Bernstein, I. D. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood 101, 1784–1789 (2003).
Karanu, F. N. et al. Human homologues of Delta-1 and Delta-4 function as mitogenic regulators of primitive human hematopoietic cells. Blood 97, 1960–1967 (2001).
Ohishi, K., Varnum-Finney, B. & Bernstein, I. D. Delta-1 enhances marrow and thymus repopulating ability of human CD34+CD38− cord blood cells. J. Clin. Invest. 110, 1165–1174 (2002).
Calvi, L. M. et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003).
Sauvageau, G. et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl Acad. Sci. USA 91, 12223–12227 (1994).
Lessard, J., Baban, S. & Sauvageau, G. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood 91, 1216–1224 (1998).
Austin, T. W., Solar, G. P., Ziegler, F. C., Liem, L. & Matthews, W. A role for the Wnt gene family in hematopoiesis: expansion of multilineage progenitor cells. Blood 89, 3624–3635 (1997).
Antonchuk, J., Sauvageau, G. & Humphries, R. K. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp. Hematol. 29, 1125–1134 (2001).
Antonchuk, J., Sauvageau, G. & Humphries, R. K. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109, 39–45 (2002).
Buske, C. et al. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood 100, 862–868 (2002).
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003).
Lessard, J. & Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 (2003).
Bhardwaj, G. et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nature Immunol. 2, 172–180 (2001).
Cheng, T. et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000).
Phillips, R. L. et al. The genetic program of hematopoietic stem cells. Science 288, 1635–1640 (2000). One of the first attempts to generate a global differential gene-expression profile of a stem-cell population compared to its progeny using subtractive hybridization of cDNA libraries.
Terskikh, A. V. et al. From hematopoiesis to neuropoiesis: evidence of overlapping genetic programs. Proc. Natl Acad. Sci. USA 98, 7934–7939 (2001).
Arai, F. et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161 (2004).
Li, L. & Akashi, K. Unraveling the molecular components and genetic blueprints of stem cells. Biotechniques 35, 1233–1239 (2003).
Rebel, V. I., Miller, C. L., Eaves, C. J. & Lansdorp, P. M. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood 87, 3500–3507 (1996).
Park, I. K. et al. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood 99, 488–498 (2002).
Liu, H. & Verfaillie, C. M. Myeloid-lymphoid initiating cells (ML-IC) are highly enriched in the rhodamine-c-kit+CD33−CD38− fraction of umbilical cord CD34+ cells. Exp. Hematol. 30, 582–589 (2002).
Hess, D. A. et al. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood 104, 1648–1655 (2004).
Gage, F. H. Mammalian neural stem cells. Science 287, 1433–1438 (2000).
Uchida, N. et al. Direct isolation of human central nervous system stem cells. Proc. Natl Acad. Sci. USA 97, 14720–14725 (2000).
Geschwind, D. H. et al. A genetic analysis of neural progenitor differentiation. Neuron 29, 325–339 (2001).
Karsten, S. L. et al. Global analysis of gene expression in neural progenitors reveals specific cell-cycle, signaling, and metabolic networks. Dev. Biol. 261, 165–182 (2003).
Wright, L. S. et al. Gene expression in human neural stem cells: effects of leukemia inhibitory factor. J. Neurochem. 86, 179–195 (2003).
Livesey, F. J., Young, T. L. & Cepko, C. L. An analysis of the gene expression program of mammalian neural progenitor cells. Proc. Natl Acad. Sci. USA 101, 1374–1379 (2004).
Luo, Y. et al. Microarray analysis of selected genes in neural stem and progenitor cells. J. Neurochem. 83, 1481–1497 (2002).
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004). Identifies two spatially distinct stem-cell populations in the bulge-cell pool and characterizes their expression profile, in vivo and in vitro potential and niche.
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nature Biotechnol. 22, 411–417 (2004).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Mills, J. C., Andersson, N., Hong, C. V., Stappenbeck, T. S. & Gordon, J. I. Molecular characterization of mouse gastric epithelial progenitor cells. Proc. Natl Acad. Sci. USA 99, 14819–14824 (2002).
Stappenbeck, T. S., Mills, J. C. & Gordon, J. I. Molecular features of adult mouse small intestinal epithelial progenitors. Proc. Natl Acad. Sci. USA 100, 1004–1009 (2003).
Gilboa, L. & Lehmann, R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development 131, 4895–4905 (2004).
Yamashita, Y. M., Fuller, M. T. & Jones, D. L. Signaling in stem cell niches: lessons from the Drosophila germline. J. Cell Sci. 118, 665–672 (2005).
Xie, T. & Spradling, A. C. A niche maintaining germ line stem cells in the Drosophila ovary. Science 290, 328–330 (2000).
Tulina, N. & Matunis, E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK–STAT signaling. Science 294, 2546–2549 (2001).
Kiger, A. A., Jones, D. L., Schulz, C., Rogers, M. B. & Fuller, M. T. Stem cell self-renewal specified by JAK–STAT activation in response to a support cell cue. Science 294, 2542–2545 (2001).
Gonczy, P. & DiNardo, S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437–24347 (1996).
Song, X., Zhu, C. H., Doan, C. & Xie, T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296, 1855–1857 (2002).
Tran, J., Brenner, T. J. & DiNardo, S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature 407, 754–757 (2000).
Deng, W. & Lin, H. Spectrosomes and fusomes anchor mitotic spindles during asymmetric germ cell divisions and facilitate the formation of a polarized microtubule array for oocyte specification in Drosophila. Dev. Biol. 189, 79–94 (1997).
Kiger, A. A., White-Cooper, H. & Fuller, M. T. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature 407, 750–754 (2000).
Zhang, J. et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841 (2003).
Shen, Q. et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science 304, 1338–1340 (2004).
Doetsch, F. A niche for adult neural stem cells. Curr. Opin. Genet. Dev. 13, 543–550 (2003).
Nishimura, E. K. et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416, 854–860 (2002).
Driessen, R. L., Johnston, H. M. & Nilsson, S. K. Membrane-bound stem cell factor is a key regulator in the initial lodgment of stem cells within the endosteal marrow region. Exp. Hematol. 31, 1284–1291 (2003).
Hackney, J. A. et al. A molecular profile of a hematopoietic stem cell niche. Proc. Natl Acad. Sci. USA 99, 13061–13066 (2002). Provides a complementary analysis of gene expression in a stem-cell niche, specifically the haematopoietic stem-cell niche, as a means to understand the extrinsic signals that regulate stem-cell function.
Yamashita, Y. M., Jones, D. L. & Fuller, M. T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550 (2003).
Reya, T. & Clevers, H. Wnt signalling in stem cells and cancer. Nature 434, 843–850 (2005).
Cobas, M. et al. β-catenin is dispensable for hematopoiesis and lymphopoiesis. J. Exp. Med. 199, 221–229 (2004).
Molofsky, A. V., Pardal, R. & Morrison, S. J. Diverse mechanisms regulate stem cell self-renewal. Curr. Opin. Cell Biol. 16, 700–707 (2004).
Espinosa, L., Ingles-Esteve, J., Aguilera, C. & Bigas, A. Phosphorylation by glycogen synthase kinase-3β down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278, 32227–32235 (2003).
Zipori, D. The nature of stem cells: state rather than entity. Nature Rev. Genet. 5, 873–878 (2004). A critical analysis of reported stem-cell gene-expression data that raises questions regarding the validity of the concept of conserved stem-cell genes.
Gygi, S. P., Rochon, Y., Franza, B. R. & Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 (1999).
Miyamoto, T. et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147 (2002).
Lian, Z. et al. Genomic and proteomic analysis of the myeloid differentiation program. Blood 98, 513–524 (2001).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Suh, M. R. et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498 (2004).
Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. MicroRNAs modulate hematopoietic lineage differentiation. Science 303, 83–86 (2004).
Pickart, M. A. et al. Functional genomics tools for the analysis of zebrafish pigment. Pigment Cell Res. 17, 461–470 (2004).
Nasevicius, A. & Ekker, S. C. Effective targeted gene 'knockdown' in zebrafish. Nature Genet. 26, 216–220 (2000).
Ekker, S. C. Morphants: a new systematic vertebrate functional genomics approach. Yeast 17, 302–306 (2000).
Heasman, J. Morpholino oligos: making sense of antisense? Dev. Biol. 243, 209–214 (2002).
Bargmann, C. I. High-throughput reverse genetics: RNAi screens in Caenorhabditis elegans. Genome Biol. 2, REVIEWS1005 (2001).
Eckfeldt, C. E. et al. Functional analysis of hematopoietic stem cell gene expressin using zebrafish. PLoS Biol. 3, e254 (2005). One of the first descriptions of global gene-expression profiling of a human stem-cell population coupled with high-throughput functional genomics screening using zebrafish.
Pomerantz, J. & Blau, H. M. Nuclear reprogramming: a key to stem cell function in regenerative medicine. Nature Cell Biol. 6, 810–816 (2004).
Grove, J. E., Bruscia, E. & Krause, D. S. Plasticity of bone marrow-derived stem cells. Stem Cells 22, 487–500 (2004).
Wagers, A. J. & Weissman, I. L. Plasticity of adult stem cells. Cell 116, 639–648 (2004).
Camargo, F. D., Chambers, S. M. & Goodell, M. A. Stem cell plasticity: from transdifferentiation to macrophage fusion. Cell Prolif. 37, 55–65 (2004).
Verfaillie, C. M. Adult stem cells: assessing the case for pluripotency. Trends Cell Biol. 12, 502–508 (2002).
Khavari, P. A. Profiling epithelial stem cells. Nature Biotechnol. 22, 393–394 (2004).
Gronthos, S. et al. Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J. Cell Sci. 116, 1827–1835 (2003).
Oh, H. et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc. Natl Acad. Sci. USA 100, 12313–12318 (2003).
Paku, S., Schnur, J., Nagy, P. & Thorgeirsson, S. S. Origin and structural evolution of the early proliferating oval cells in rat liver. Am. J. Pathol. 158, 1313–1323 (2001).
Petkov, P. M. et al. Gene expression pattern in hepatic stem/progenitor cells during rat fetal development using complementary DNA microarrays. Hepatology 39, 617–627 (2004).
Arai, M. et al. Gene expression profiles in liver regeneration with oval cell induction. Biochem. Biophys. Res. Comm. 317, 370–376 (2004).
Seaberg, R. M. et al. Clonal identification of multipotent precursors from adult mouse pancreas that generate neural and pancreatic lineages. Nature Biotechnol. 22, 1115–1124 (2004).
Bonner-Weir, S. et al. The pancreatic ductal epithelium serves as a potential pool of progenitor cells. Pediatr. Diabetes 5, 16–22 (2004).
Hawke, T. J. & Garry, D. J. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 91, 534–551 (2001).
de Rooij, D. G. & Grootegoed, J. A. Spermatogonial stem cells. Curr. Opin. Cell Biol. 10, 694–701 (1998).
Acknowledgements
The authors would like to acknowledge the work of our colleagues that we have discussed, and apologize to our colleagues whose work was not discussed due to space limitations.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Gene
FURTHER INFORMATION
Catherine Verfaillie's Institute
National Institutes of Health Stem Cell Information
Glossary
- PLURIPOTENT
-
The ability to give rise to all embryonic tissues, but not extra-embryonic tissues. Totipotent cells can give rise to all embryonic and extra-embryonic (trophectodermal) tissues.
- INNER CELL MASS
-
A mass of pluripotent cells in the interior of the developing blastocyst that give rise to all embryonic tissues. The blastocyst is part of the pre-implantation-stage embryo and consists of a hollow sphere of cells with a distinct outer trophectoderm layer and an inner cell mass.
- MULTIPOTENT
-
The ability to give rise to the diverse cell types of one or a few tissues.
- GENITAL RIDGE
-
The bilateral structures in the developing embryo that give rise to the gonads.
- TRANSCRIPTOME
-
The entire transcriptional repertoire of a cell or cell population.
- TROPHECTODERM
-
The outer portion of the blastocyst that gives rise to the embryonic portion of the placenta.
- HOMEODOMAIN TRANSCRIPTION FACTOR
-
A transcription factor that contains a homeodomain DNA-binding domain.
- EST SEQUENCING
-
The sequencing of short segments of expressed genes (expressed sequence tags or ESTs) present in cDNA libraries that can be used for gene cloning or to show which genes are present in a cell population.
- TRANS-DIFFERENTIATE
-
Differentiation of one cell type directly to another cell type without dedifferentiation to a more primitive intermediate.
- STEM-CELL PLASTICITY
-
The apparent ability of a stem/progenitor cell fated to a particular tissue to acquire a differentiated phenotype of a different tissue.
- cRNA
-
A complementary RNA molecule that hybridizes with a specific messenger RNA sequence.
- IN SILICO DIFFERENTIAL DISPLAY
-
The use of computer algorithms to determine differential expression of transcripts from gene-expression databases.
- FLUORESCENCE ACTIVATED CELL SORTING
-
Automated, high-speed sorting of cell populations based on the presence of intrinsic fluorescent labels such as GFP expression, or extrinsic fluorescent labels such as monoclonal antibodies conjugated to fluorochromes.
- HOMEOBOX (HOX) GENE FAMILY
-
A family of transcriptional regulators that share a conserved homeobox DNA-binding domain, and that are involved in the regulation of embryonic and adult developmental fates.
- POLYCOMB (PCG) GENE FAMILY
-
Genes encoding a family of proteins that form complexes that modify chromatin structure and selectively repress gene transcription.
- WNT GENE FAMILY
-
A family of genes that mediate intercellular signalling through secreted glycoprotein Wnt ligands.
- microRNA
-
A family of short, non-coding RNA molecules (∼22 nucleotides) that post-transcriptionally regulate target-gene expression primarily by inhibiting protein translation.
- RNAi
-
A functional tool that use small interfering RNAs (siRNAs) to knock down gene expression through sequence-specific decay of target mRNA molecules.
- MORPHOLINO ANTISENSE OLIGONUCLEOTIDES
-
Chemically synthesized oligonucleotide analogues used to knock down gene expression by specifically binding to target transcripts to inhibit RNA splicing or translation.
- CHIP
-
Technique used to immunoprecipitate complexes of DNA with associated proteins.
Rights and permissions
About this article
Cite this article
Eckfeldt, C., Mendenhall, E. & Verfaillie, C. The molecular repertoire of the 'almighty' stem cell. Nat Rev Mol Cell Biol 6, 726–737 (2005). https://doi.org/10.1038/nrm1713
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1713
This article is cited by
-
Reduction, proliferation, and differentiation defects of stem cells over time: a consequence of selective accumulation of old centrioles in the stem cells?
Molecular Biology Reports (2023)
-
Can Extracellular Vesicles as Drug Delivery Systems Be a Game Changer in Cardiac Disease?
Pharmaceutical Research (2023)
-
Prostaglandin E2 Regulates Bipotent Monocyte-Dendritic Progenitor Cell Lineage-Commitment
Stem Cell Reviews and Reports (2021)
-
Synchrotron- and focal plane array-based Fourier-transform infrared spectroscopy differentiates the basalis and functionalis epithelial endometrial regions and identifies putative stem cell regions of human endometrial glands
Analytical and Bioanalytical Chemistry (2018)
-
Isolation, culture, characterization, and adipogenic differentiation of heifer endometrial mesenchymal stem cells
Comparative Clinical Pathology (2015)