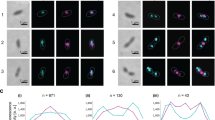
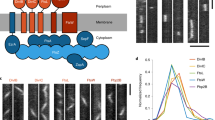
Key Points
-
FtsZ is required for the formation of a ring structure, known as the Z ring, at the site of cell or organelle division. Contraction of the Z ring leads to fission of the cell or organelle.
-
The FtsZ family of proteins is widely conserved among prokaryotes and plants where it is used in chloroplast division. However, FtsZ is notably absent in a branch of the archaea, several branches of bacteria and in the mitochondria of most eukaryotes.
-
FtsZ is a structural homologue of tubulin, and its biochemical properties also resemble those of tubulins, including its ability to bind and hydrolyse GTP and assemble into protofilaments. Unlike tubulin, however, FtsZ does not form microtubules, and the precise nature of FtsZ-containing macromolecules in vivo is not known.
-
The Z ring localizes to the centre of Escherichia coli cells at the correct time during the cell cycle. There has been much progress in understanding how two negative regulatory systems ensure the proper spatial regulation of the Z ring. Several regulators of Z-ring assembly have been discovered that might interact with the spatial and temporal regulatory network.
-
Whereas the requirement of FtsZ in cytokinesis is well-established, it is unclear whether the Z ring drives cytokinesis actively by mechanical pulling against the membrane or more passively by the recruitment of cell-wall-synthesis enzymes, or both. Recent evidence indicates that a significant proportion of the FtsZ protein population is not ring associated, and that these proteins might have additional functions besides cytokinesis, such as the maintenance of cell shape.
-
Good progress has been made in understanding the important role of FtsZ in the fission of organelles, particularly in chloroplasts. FtsZ proteins work along with dynamin and other components to divide organelles.
Abstract
Binary fission of many prokaryotes as well as some eukaryotic organelles depends on the FtsZ protein, which self-assembles into a membrane-associated ring structure early in the division process. FtsZ is homologous to tubulin, the building block of the microtubule cytoskeleton in eukaryotes. Recent advances in genomics and cell-imaging techniques have paved the way for the remarkable progress in our understanding of fission in bacteria and organelles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bi, E. & Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991).
Weiss, D. S. Bacterial cell division and the septal ring. Mol. Microbiol. 54, 588–597 (2004).
Vaughan, S., Wickstead, B., Gull, K. & Addinall, S. G. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 58, 19–29 (2004).
Sontag, C. A., Staley, J. T. & Erickson, H. P. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J. Cell Biol. 169, 233–238 (2005). A member of the Chlamydia/Verrucomicrobia group of bacteria that lacks ftsZ contains instead two genes that are evolutionarily closer to tubulin than FtsZ. This paper investigates the assembly and nucleotide-binding properties of these two bacterial tubulins.
Glass, J. I. et al. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407, 757–762 (2000).
Osteryoung, K. W. & Nunnari, J. The division of endosymbiotic organelles. Science 302, 1698–1704 (2003). This is a recent comprehensive review of organelle division.
Stokes, K. D. & Osteryoung, K. W. Early divergence of the FtsZ1 and FtsZ2 plastid division gene families in photosynthetic eukaryotes. Gene 320, 97–108 (2003).
Miyagishima, S. Y., Nozaki, H., Nishida, K., Matsuzaki, M. & Kuroiwa, T. Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: the duplication of FtsZ is implicated in endosymbiosis. J. Mol. Evol. 58, 291–303 (2004).
Löwe, J. & Amos, L. A. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 (1998).
Oliva, M. A., Cordell, S. C. & Löwe, J. Structural insights into FtsZ protofilament formation. Nature Struct. Mol. Biol. 11, 1243–1250 (2004). The core domain of FtsZ consists of independently folding N-terminal and C-terminal domains. In addition, this study and reference 28 suggest that nucleotide exchange occurs rapidly, with hydrolysis being the limiting step.
Ma, X. & Margolin, W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181, 7531–7544 (1999).
Pichoff, S. & Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693 (2002).
Redick, S. D., Stricker, J., Briscoe, G. & Erickson, H. P. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J. Bacteriol. 187, 2727–2236 (2005).
Mukherjee, A., Dai, K. & Lutkenhaus, J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl Acad. Sci. USA 90, 1053–1057 (1993).
de Boer, P., Crossley, R. & Rothfield, L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254–256 (1992).
Raychaudhuri, D. & Park, J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254 (1992).
Mukherjee, A. & Lutkenhaus, J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176, 2754–2758 (1994).
Erickson, H. P., Taylor, D. W., Taylor, K. A. & Bramhill, D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. USA 93, 519–523 (1996).
Löwe, J. & Amos, L. A. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18, 2364–2371 (1999).
Erickson, H. P. & Stoffler, D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to α, β, and γ tubulin. J. Cell Biol. 135, 5–8 (1996).
Yu, X. -C. & Margolin, W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 16, 5455–5463 (1997).
Gonzalez, J. M. et al. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 278, 37664–37671 (2003).
Raychaudhuri, D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18, 2372–2383 (1999).
White, E. L. et al. Slow polymerization of Mycobacterium tuberculosis FtsZ. J. Bacteriol. 182, 4028–4034 (2000).
Sossong, T. M. Jr, Brigham-Burke, M. R., Hensley, P. & Pearce, K. H. Jr. Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry 38, 14843–14850 (1999).
Scheffers, D. J., de Wit, J. G., den Blaauwen, T. & Driessen, A. J. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41, 521–529 (2002).
Romberg, L. & Levin, P. A. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57, 125–154 (2003).
Romberg, L. & Mitchison, T. J. Rate-limiting guanosine 5´-triphosphate hydrolysis during nucleotide turnover by FtsZ, a prokaryotic tubulin homologue involved in bacterial cell division. Biochemistry 43, 282–288 (2004).
Mukherjee, A. & Lutkenhaus, J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 (1998).
Desai, A. & Mitchison, T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117 (1997).
Huecas, S. & Andreu, J. M. Energetics of the cooperative assembly of cell division protein FtsZ and the nucleotide hydrolysis switch. J. Biol. Chem. 278, 46146–46154 (2003).
Lu, C., Reedy, M. & Erickson, H. P. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182, 164–170 (2000).
Caplan, M. & Erickson, H. P. Apparent cooperative assembly of the bacterial cell-division protein FtsZ demonstrated by isothermal titration calorimetry. J. Biol. Chem. 278, 13784–13788 (2003).
Chen, Y., Bjornson, K., Redick, S. D. & Erickson, H. P. A rapid fluorescence assay for FtsZ assembly indicates cooperative assembly with a dimer nucleus. Biophys. J. 88, 505–514 (2005).
Rivas, G. et al. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. J. Biol. Chem. 275, 11740–11749 (2000).
Romberg, L., Simon, M. & Erickson, H. P. Polymerization of FtsZ, a bacterial homolog of tubulin: Is assembly cooperative? J. Biol. Chem. 276, 11743–11753 (2001).
Gonzalez, J. M. et al. Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc. Natl Acad. Sci. USA 102, 1895–1900 (2005).
Chen, Y. & Erickson, H. P. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 280, 22549–22554 (2005).
Anderson, D. E., Gueiros-Filho, F. J. & Erickson, H. P. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186, 5775–5781 (2004). Although the Z ring seems to be stable, FtsZ subunits turn over rapidly within the ring, with a half-time of 8–9 s.
Raskin, D. M. & de Boer, P. A. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA 96, 4971–4976 (1999).
Hu, Z., Mukherjee, A., Pichoff, S. & Lutkenhaus, J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA 96, 14819–14824 (1999).
Suefuji, K., Valluzzi, R. & Raychaudhuri, D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl Acad. Sci. USA 99, 16776–16781 (2002).
Hu, Z. & Lutkenhaus, J. Topological regulation of cell division in E. coli.: spatiotemporal scillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7, 1337–1343 (2001).
Huang, K. C., Meir, Y. & Wingreen, N. S. Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc. Natl Acad. Sci. USA 100, 12724–12728 (2003).
Thanedar, S. & Margolin, W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14, 1167–1173 (2004). Non-ring FtsZ is not randomly dispersed within the cell but instead oscillates from pole to pole in a Min-dependent manner, in a spiral pattern.
Marston, A. L., Thomaides, H. B., Edwards, D. H., Sharpe, M. E. & Errington, J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12, 3419–3430 (1998).
Migocki, M. D., Freeman, M. K., Wake, R. G. & Harry, E. J. The Min system is not required for precise placement of the midcell Z ring in Bacillus subtilis. EMBO Rep. 3, 1163–1167 (2002).
Szeto, J. et al. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183, 6253–6264 (2001).
Mazouni, K., Domain, F., Cassier-Chauvat, C. & Chauvat, F. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52, 1145–1158 (2004).
Corbin, B. D., Yu, X. -C. & Margolin, W. Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J. 21, 1988–2008 (2002).
Ramirez-Arcos, S., Szeto, J., Dillon, J. A. & Margolin, W. Conservation of dynamic localization among MinD and MinE orthologues: oscillation of Neisseria gonorrhoeae proteins in Escherichia coli. Mol. Microbiol. 46, 493–504 (2002).
Woldringh, C. et al. Role of the nucleoid in the toporegulation of division. Res. Microbiol. 141, 39–49 (1990).
Sun, Q. & Margolin, W. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183, 1413–1422 (2001).
Yu, X. -C. & Margolin, W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32, 315–326 (1999).
Quardokus, E. M. & Brun, Y. V. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol. Microbiol. 45, 605–616 (2002).
Sun, Q. & Margolin, W. Effects of perturbing nucleoid structure on nucleoid occlusion-mediated toporegulation of FtsZ ring assembly. J. Bacteriol. 186, 3951–3959 (2004).
Bernhardt, T. G. & de Boer, P. A. SlmA, a nucleoid-associated, FtsZ-binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 (2005). This paper, along with reference 58, demonstrates that nucleoid occlusion is mediated by DNA-binding proteins that interact with FtsZ.
Wu, L. J. & Errington, J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915–925 (2004).
Janakiraman, A. & Goldberg, M. B. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl Acad. Sci. USA 101, 835–840 (2004).
Mileykovskaya, E. & Dowhan, W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 182, 1172–1175 (2000).
Miyagishima, S. Y., Wolk, C. P. & Osteryoung, K. W. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 56, 126–143 (2005).
Ramos, A. et al. Altered morphology produced by ftsZ expression in Corynebacterium glutamicum ATCC 13869. Microbiology 151, 2563–2572 (2005).
Den Blaauwen, T., Buddelmeijer, N., Aarsman, M. E., Hameete, C. M. & Nanninga, N. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181, 5167–5175 (1999).
Gullbrand, B. & Nordström, K. FtsZ ring formation without subsequent cell division after replication runout in Escherichia coli. Mol. Microbiol. 36, 1349–1359 (2000).
Harry, E. J., Rodwell, J. & Wake, R. G. Co-ordinating DNA replication with cell division in bacteria: a link between the early stages of a round of replication and mid-cell Z ring assembly. Mol. Microbiol. 33, 33–40 (1999).
Migocki, M. D., Lewis, P. J., Wake, R. G. & Harry, E. J. The midcell replication factory in Bacillus subtilis is highly mobile: implications for coordinating chromosome replication with other cell cycle events. Mol. Microbiol. 54, 452–463 (2004).
de Pedro, M. A., Quintela, J. C., Holtje, J. -V. & Schwarz, H. Murein segregation in Escherichia coli. J. Bacteriol. 179, 2823–2834 (1997).
Bach, T. & Skarstad, K. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol. Microbiol. 51, 1589–1600 (2004).
Liu, G., Begg, K., Geddes, A. & Donachie, W. D. Transcription of essential cell division genes is linked to chromosome replication in Escherichia coli. Mol. Microbiol. 40, 909–916 (2001).
Weart, R. B. & Levin, P. A. Growth rate-dependent regulation of medial FtsZ ring formation. J. Bacteriol. 185, 2826–2834 (2003).
Kelly, A. J., Sackett, M., Din, N., Quardokus, E. & Brun, Y. V. Cell cycle dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 12, 880–893 (1998).
Quardokus, E. M., Din, N. & Brun, Y. V. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39, 949–959 (2001).
Hale, C. A. & de Boer, P. A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997).
Levin, P. A., Kurtser, I. G. & Grossman, A. D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl Acad. Sci. USA 96, 9642–9647 (1999).
Haeusser, D. P., Schwartz, R. L., Smith, A. M., Oates, M. E. & Levin, P. A. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52, 801–814 (2004).
Weart, R. B., Nakano, S., Lane, B. E., Zuber, P. & Levin, P. A. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57, 238–249 (2005).
Low, H. H., Moncrieffe, M. C. & Lowe, J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341, 839–852 (2004).
Gueiros-Filho, F. J. & Losick, R. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 (2002).
Johnson, J. E., Lackner, L. L., Hale, C. A. & de Boer, P. A. ZipA is required for targeting of DMinC/DicB, but not DMinC/MinD, complexes to septal ring assemblies in Escherichia coli. J. Bacteriol. 186, 2418–2429 (2004).
Huisman, O. & D'Ari, R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature 290, 797–799 (1981).
Bi, E. & Lutkenhaus, J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175, 1118–1125 (1993).
Cordell, S. C., Robinson, E. J. & Lowe, J. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl Acad. Sci. USA 100, 7889–7894 (2003).
Kawai, Y., Moriya, S. & Ogasawara, N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol. Microbiol. 47, 1113–1122 (2003).
Pichoff, S. & Lutkenhaus, J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55, 1722–1734 (2005). An independent membrane-targeting sequence is required for FtsA to interact with the membrane, which in turn helps to keep the Z ring anchored to the membrane.
Geissler, B., Elraheb, D. & Margolin, W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 4197–4202 (2003).
Datta, P., Dasgupta, A., Bhakta, S. & Basu, J. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 277, 24983–24987 (2002).
Buddelmeijer, N. & Beckwith, J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52, 1315–1327 (2004).
Goehring, N. W., Gueiros-Filho, F. & Beckwith, J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 19, 127–137 (2005).
Corbin, B. D., Geissler, B., Sadasivam, M. & Margolin, W. A Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186, 7736–7744 (2004). This paper, along with reference 88, uses novel cytological methods for dissecting the complex interactions among cell-division proteins and provides additional evidence for the existence of membrane-protein subassemblies.
Di Lallo, G., Fagioli, M., Barionovi, D., Ghelardini, P. & Paolozzi, L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149, 3353–3359 (2003).
Karimova, G., Dautin, N. & Ladant, D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187, 2233–2243 (2005).
Robson, S. A., Michie, K. A., Mackay, J. P., Harry, E. J. & King, G. F. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol. Microbiol. 44, 663–674 (2002).
Martin, M. E., Trimble, M. J. & Brun, Y. V. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 54, 60–74 (2004).
Pinho, M. G. & Errington, J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50, 871–881 (2003).
Morlot, C., Noirclerc-Savoye, M., Zapun, A., Dideberg, O. & Vernet, T. The D, D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51, 1641–1648 (2004).
Pinho, M. G. & Errington, J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55, 799–807 (2005).
Praefcke, G. J. & McMahon, H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature Rev. Mol. Cell Biol. 5, 133–147 (2004).
Sun, Q. & Margolin, W. FtsZ dynamics during the cell division cycle of live Escherichia coli. J. Bacteriol. 180, 2050–2056 (1998).
Figge, R. M., Divakaruni, A. V. & Gober, J. W. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51, 1321–1332 (2004).
Varma, A. & Young, K. D. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 186, 6768–6774 (2004). FtsZ is involved in the maintenance of proper cell shape in addition to its established role in cytokinesis.
Osteryoung, K. W., Stokes, K. D., Rutherford, S. M., Percival, A. L. & Lee, W. Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial FtsZ. Plant Cell 10, 1991–2004 (1998).
Miyagishima, S., Takahara, M. & Kuroiwa, T. Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 13, 707–721 (2001).
Miyagishima, S. Y. et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15, 655–665 (2003).
Gao, H., Kadirjan-Kalbach, D., Froehlich, J. E. & Osteryoung, K. W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl Acad. Sci. USA 100, 4328–4333 (2003).
Thompson, H. M., Skop, A. R., Euteneuer, U., Meyer, B. J. & McNiven, M. A. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr. Biol. 12, 2111–2117 (2002).
Kang, B. H., Busse, J. S. & Bednarek, S. Y. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913 (2003).
Vitha, S., McAndrew, R. S. & Osteryoung, K. W. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153, 111–120 (2001).
Colletti, K. S. et al. A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10, 507–516 (2000).
Itoh, R., Fujiwara, M., Nagata, N. & Yoshida, S. A chloroplast protein homologous to the eubacterial topological specificity factor MinE plays a role in chloroplast division. Plant Physiol. 127, 1644–1655 (2001).
Vitha, S. et al. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15, 1918–1933 (2003). Evidence is mounting that several cyanobacterial cell-division proteins might have been retained by plants for chloroplast division.
Raynaud, C., Cassier-Chauvat, C., Perennes, C. & Bergounioux, C. An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16, 1801–1811 (2004).
Maple, J. et al. GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr. Biol. 14, 776–781 (2004).
Fulgosi, H., Gerdes, L., Westphal, S., Glockmann, C. & Soll, J. Cell and chloroplast division requires ARTEMIS. Proc. Natl Acad. Sci. USA 99, 11501–11506 (2002).
Fadda, D. et al. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J. Bacteriol. 185, 6209–6214 (2003).
Reski, R. Rings and networks: the amazing complexity of FtsZ in chloroplasts. Trends Plant Sci. 7, 103–105 (2002).
Kiessling, J. et al. Dual targeting of plastid division protein FtsZ to chloroplasts and the cytoplasm. EMBO Rep. 5, 889–894 (2004).
Nishida, K. et al. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl Acad. Sci. USA 100, 2146–2151 (2003).
Bleazard, W. et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nature Cell Biol. 1, 298–304 (1999).
Kiefel, B. R., Gilson, P. R. & Beech, P. L. Diverse eukaryotes have retained mitochondrial homologues of the bacterial division protein FtsZ. Protist 155, 105–115 (2004).
Beech, P. L. et al. Mitochondrial FtsZ in a chromophyte alga. Science 287, 1276–1279 (2000).
Gilson, P. R. et al. Two Dictyostelium orthologs of the prokaryotic cell division protein FtsZ localize to mitochondria and are required for the maintenance of normal mitochondrial morphology. Eukaryot. Cell 2, 1315–1326 (2003).
Acknowledgements
Work in the Margolin laboratory is supported by grants from the National Institutes of Health and the National Science Foundation. I thank H. Erickson for helpful advice. I apologize to colleagues whose work was not cited here because of space limitations.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Glossary
- EURYARCHAEAL
-
Pertaining to the group of archaea that includes the methanogens and extreme halophiles.
- CRENARCHAEA
-
The group of archaea that includes the extreme thermophiles.
- NUCLEOID
-
The organized form of a bacterial chromosome.
- MYCOPLASMAS
-
Wall-less bacteria.
- ENDOSYMBIOTIC
-
Describing the engulfment of one cell by another larger cell, with the engulfed cell evolving into an organelle.
- COOPERATIVE ASSEMBLY
-
The affinity of subunits for a polymer increases as more subunits are assembled, displaying a critical concentration below which little assembly occurs.
- ISODESMIC ASSEMBLY
-
The opposite of cooperative assembly, in that the affinity of each new subunit for a polymer is independent of the subunit concentration.
- MIN SYSTEM
-
A group of two or three bacterial proteins that inhibit unwanted formation of the Z ring at the cell poles.
- REPLISOME
-
The DNA-replication protein machinery.
- SOS RESPONSE
-
Inducible DNA repair system in bacteria invoked in response to a sudden increase in DNA damage.
- BITOPIC
-
Describes an integral membrane protein that has one cytoplasmic, transmembrane and periplasmic domain.
- FRAP
-
(Fluorescence recovery after photobleaching). A microscopic technique used to measure the movement (for example, diffusion rates) of fluorescently tagged molecules over time in vivo. Specific regions in a cell are irreversibly photobleached using a laser; fluorescence is restored by diffusion of fluorescently tagged unbleached molecules into the bleached area.
- DYNAMINS
-
A family of GTPases that are important for membrane scission.
- STROMAL COMPARTMENT
-
The inner compartment of the chloroplast.
Rights and permissions
About this article
Cite this article
Margolin, W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6, 862–871 (2005). https://doi.org/10.1038/nrm1745
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm1745
This article is cited by
-
Wolbachia supergroup A in Enoplognatha latimana (Araneae: Theridiidae) in Poland as an example of possible horizontal transfer of bacteria
Scientific Reports (2024)
-
Mycobacterial FtsZ and inhibitors: a promising target for the anti-tubercular drug development
Molecular Diversity (2023)
-
Proteolysis dependent cell cycle regulation in Caulobacter crescentus
Cell Division (2022)
-
RNA-cleaving DNAzymes as a diagnostic and therapeutic agent against antimicrobial resistant bacteria
Current Genetics (2022)
-
The lack of the cell division protein FtsZ induced generation of giant cells under acidic stress in cyanobacterium Synechocystis sp. PCC6803
Photosynthesis Research (2021)