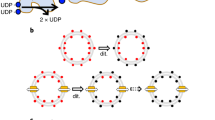
Key Points
-
Iron is one of the most important nutrients of bacteria, because of its essential metabolic role. However, iron is scarcely available under physiological conditions; first, because of its propensity to form insoluble complexes, and second, as a result of the existence of numerous iron-binding proteins that the host itself uses to store and transport iron.
-
Bacteria have evolved a wide range of strategies to overcome iron shortage and to ensure sufficient uptake. One of these relies on the synthesis and excretion of siderophores, or small compounds that either bind free iron or sequester it from iron-binding proteins in the bacterial environment.
-
In Gram-negative bacteria, recovery of iron-loaded siderophores involves a sophisticated uptake mechanism. At the outer-membrane level, high-affinity receptors capture siderophores and mediate their translocation into the periplasmic space. This is powered by the Ton complex, which resides in cytoplasmic membrane and is coupled to the proton-motive force. Siderophores are subsequently transported across the periplasm and the cytoplasmic membrane by periplasmic-binding proteins and ATP-dependent membrane transporters.
-
In recent years important advancements have been made in the characterization of the siderophore uptake system, owing to the combination of structural, biophysical, biochemical and genetic approaches. Among these, it is worth noting the determination of the atomic structure of several outer-membrane receptors and the use of spectroscopic techniques to monitor the uptake process in vivo.
-
However, fundamental questions concerning almost every aspect of the uptake process remain unclear and are the subject of continued debate between researchers.
Abstract
The outer membrane of Gram-negative bacteria constitutes a permeability barrier that protects the cell from exterior hazards, but also complicates the uptake of nutrients. In the case of iron, the challenge is even greater, because of the scarcity of this indispensable element in the cell's surroundings. To solve this dilemma, bacteria have evolved sophisticated mechanisms whereby the concerted actions of receptor, transporter and energy-transducing proteins ensure that there is a sufficient supply of iron-containing compounds, such as siderophores.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ratledge, C. & Dover, L. G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941 (2000).
Crichton, R. R. Inorganic biochemistry of iron metabolism: from molecular mechanisms to clinical consequences (John Wiley & Sons, New York, 2001).
Braun, V. & Killmann, H. Bacterial solutions to the iron-supply problem. Trends Biochem. Sci. 24, 104–109 (1999).
Clarke, T. E., Tari, L. W. & Vogel, H. J. Structural biology of bacterial iron uptake systems. Curr. Top. Med. Chem. 1, 7–30 (2001).
Kadner, R. J. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol. Microbiol. 4, 2027–2033 (1990). This review, together with references 6 and 11 – 14, illustrates the evolution of the field of TonB-dependent transport during the past decade. It describes the various permeation mechanisms that were proposed before the determination of the structure of the outer-membrane receptors.
Postle, K. TonB and the Gram-negative dilemma. Mol. Microbiol. 4, 2019–2025 (1990).
Nikaido, H. in Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt, F.) 29–47 (American Society for Microbiology, Washington DC, 1996).
Beveridge, T. J. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 (1999).
Koebnik, R., Locher, K. P. & van Gelder, P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37, 239–253 (2000).
Bradbeer, C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175, 3146–3150 (1993).
Postle, K. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 25, 591–601 (1993).
Klebba, P. E., Rutz, J. M., Liu, J. & Murphy, C. K. Mechanisms of TonB-catalyzed iron transport through the enteric bacterial cell envelope. J. Bioenerg. Biomembr. 25, 603–611 (1993).
Braun, V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16, 295–307 (1995).
Moeck, G. & Coulton, J. W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28, 675–681 (1998).
Buchanan, S. K. β-Barrel proteins from bacterial outer membranes: structure, function and refolding. Curr. Opin. Struct. Biol. 9, 455–461 (1999).
Schulz, G. E. β-barrel membrane proteins. Curr. Opin. Struct. Biol. 10, 443–447 (2000).
Postle, K. Close before opening. Science 295, 1658–1659 (2002).
Cowan, S. W. et al. Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733 (1992).
Schirmer, T., Keller, T. A., Wang, Y. F. & Rosenbusch, J. P. Structural basis for sugar translocation through maltoporin channels at 3.1Å resolution. Science 267, 512–514 (1995).
Klebba, P. E. & Newton, S. M. C. Mechanisms of solute transport through the outer membrane proteins: burning down the house. Curr. Biol. 1, 238–248 (1998).
Schirmer, T. General and specific porins from bacterial outer membranes. J. Struct. Biol. 121, 101–109 (1998).
Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K. & Welte, W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282, 2215–2220 (1998).
Locher, K. P. et al. Transmembrane signalling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95, 771–778 (1998). References 22 and 23 reported simultaneously the atomic structures of the ferrichrome receptor and transporter FhuA in the ligand-free and -bound states, revealing for the first time the existence of an additional protein domain that blocked the permeation pathway, and showing its role in signalling.
Buchanan, S. K. et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nature Struct. Biol. 6, 56–63 (1999). This paper reported the atomic structure of the enterobactin receptor and transporter FepA, verifying the common protein architecture of TonB-dependent receptors.
Ferguson, A. D. et al. Structural basis of gating by the outer membrane transporter FecA. Science 295, 1715–1719 (2002). This paper reported the atomic structure of FecA in the ligand-free and -bound states, revealing allosteric changes in the extracellular domain that seem to correspond to a gating mechanism, in addition to changes that involved the plug domain as reported previously.
Scott, D. C. et al. Exchangeability of N-termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 276, 13025–13033 (2001).
Moeck, G., Coulton, J. W. & Postle, K. Cell envelope signalling in Escherichia coli: ligand binding to the ferrichrome-iron receptor FhuA promotes interaction with the energy-transducing protein TonB. J. Biol. Chem. 272, 28391–28397 (1997).
Braun, V. Pumping iron through cell membranes. Science 282, 2202–2203 (1998).
Tuckman, M. & Osburne, M. S. In vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. J. Bacteriol. 174, 320–323 (1992).
Moeck, G. et al. Ligand-induced conformational change in the ferrichrome-iron receptor of Escherichia coli. Mol. Microbiol. 22, 459–471 (1996).
Larsen, R. A., Foster-Hartnett, D., McIntosh, M. A. & Postle, K. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179, 3213–3221 (1997).
Cadieux, N., Bradbeer, C. & Kadner, R. J. Sequence changes in the TonB box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182, 5954–5961 (2000).
Barnard, T. J., Watson, M. E. & McIntosh, M. A. Mutations in Escherichia coli receptor FepA reveal residues involved in ligand binding and transport. Mol. Microbiol. 41, 527–536 (2001).
Merianos, H. J., Cadieux, N., Lin, C. H., Kadner, R. J. & Cafiso, D. S. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nature Struct. Biol. 7, 205–209 (2000).
Coggshall, K. A., Cadieux, N., Piedmont, C., Kadner, R. J. & Cafiso, D. S. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry 40, 13964–13971 (2001).
Chimento, D. P., Mohanty, A. K., Kadner, R. J. & Wiener, M. C. Expression, purification, characterization and crystallization of the E. coli outer membrane cyanocobalamin transporter BtuB. Biophys. J. 82, 2754A (2002).
Wiener, M. C., Chimento, D. P., Mohanty, A. K. & Kadner, R. J. The crystal structure of the E. coli outer membrane cyanocobalamin transporter BtuB. Biophys. J. 82, 2514A (2002).
Braun, M., Killman, H. & Braun, V. The β-barrel domain of FhuAΔ5-160 is sufficient for TonB-dependent activities of Escherichia coli. Mol. Microbiol. 33, 1037–1049 (1999). This paper, together with references 26 and 40, is concerned with the activity of receptors that either lack the plug domain or contain a non-native homologue, which illustrates the current debate about the functional relevance of this domain.
Bonhivers, M. et al. Stability studies of FhuA, a two-domain outer membrane protein from Escherichia coli. Biochemistry 40, 2606–2613 (2001).
Vakharia, H. & Postle, K. FepA with globular domain deletions lacks activity. J. Bacteriol. 184, 5508–5512 (2002).
Liu, J., Rutz, J. M., Klebba, P. E. & Feix, J. B. A site-directed spin-labeling study of ligand–induced conformational change in the ferric enterobactin receptor, FepA. Biochemistry 33, 13274–13283 (1994).
Jiang, X. Q. et al. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science 276, 1261–1264 (1997).
Bös, C., Lorenzen, D. & Braun, V. Specific in vivo labeling of cell surface-exposed protein loops: reactive cysteines in the predicted gating loop mark a ferrichrome binding site and a ligand-induced conformational change of the Escherichia coli FhuA protein. J. Bacteriol. 180, 605–613 (1998).
Klug, C. S., Eaton, S. S., Eaton, G. R. & Feix, J. B. Ligand-induced conformational change in the ferric enterobactin receptor FepA as studied by site-directed spin labeling and time-domain ESR. Biochemistry 37, 9016–9023 (1998).
Scott, D. C., Newton, S. M. C. & Klebba, P. E. Surface loop motion in FepA. J. Bacteriol. 184, 4906–4911 (2002).
Faraldo-Gómez, J. D., Smith, G. R. & Sansom, M. S. P. Molecular dynamics simulations of the bacterial outer membrane protein FhuA: a study of the ferrichrome-free and bound states. Biophys. J. (in the press).
Karplus, M. & Petsko, G. A. Molecular dynamics simulations in biology. Nature 347, 631–639 (1990).
Hansson, T., Oostenbrink, C. & Van Gunsteren, W. F. Molecular dynamics simulations. Curr. Opin. Struct. Biol. 12, 190–196 (2002).
Karplus, M. & McCammon, J. A. Molecular dynamics simulations of biomolecules. Nature Struct. Biol. 9, 646–652 (2002).
Folschweiller, N. et al. The pyoverdin receptor FpvA, a TonB-dependent receptor involved in iron uptake by Pseudomonas aeruginosa. Mol. Membr. Biol. 17, 123–133 (2000).
Schalk, I. J. et al. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: implications for siderophore-mediated iron transport. Biochemistry 38, 9357–9365 (1999).
Schalk, I. J. et al. Iron-free pyoverdin binds to its outer membrane receptor FpvA in Pseudomonas aeruginosa: a new mechanism for membrane iron transport. Mol. Microbiol. 39, 351–360 (2001).
Schalk, I. J., Abdallah, M. A. & Pattus, F. Recycling of pyoverdin on the FpvA receptor after ferric pyoverdin uptake and dissociation in Pseudomonas aeruginosa. Biochemistry 41, 1663–1671 (2002). In the studies reported in references 51 – 53, FRET and radiolabelling techniques were used to characterize the association of the receptor FpvA with the siderophore pyoverdin, as well as to monitor its uptake and recycling into the medium.
Stintzi, A., Barnes, C., Jide, X. & Raymond, K. N. Microbial iron-transport via a siderophore shuttle: a membrane ion transport paradigm. Proc. Natl Acad. Sci. USA 97, 10691–10696 (2000).
Braun, V. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol. Chem. 378, 779–786 (1997).
Escolar, L., Pérez-Martín, J. & de Lorenzo, V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229 (1999).
Braun, V. Surface signalling: novel transcription initiation mechanism starting from the cell surface. Arch. Microbiol. 167 (1997). This review and reference 59 describe the process whereby the transcription of the Fec uptake system is regulated by the presence of ferric citrate at the level of the cell surface.
Angerer, A. & Braun, V. Iron regulates transcription of the Escherichia coli ferric citrate transport genes directly and through the transcription initiation proteins. Arch. Microbiol. 169, 483–490 (1998).
Enz, S., Mahren, S., Stroeher, U. W. & Braun, V. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR and FecI regulatory proteins. J. Bacteriol. 182, 637–646 (2000).
Stiefel, A. et al. Control of the ferric citrate transport system of Escherichia coli: mutations in region 2.1 of the FecI extracytoplasmic-function sigma factor suppress mutations in the FecR transmembrane regulatory protein. J. Bacteriol. 183, 162–170 (2001).
Kim, I., Stiefel, A., Plantör, A., Angerer, A. & Braun, V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23, 333–344 (1997).
Howard, S. P., Herrmann, C., Stratilo, C. W. & Braun, V. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J. Bacteriol. 183, 5885–5895 (2001).
Wang, C. & Newton, A. An additional step in the transport of iron defined by the tonb locus of Escherichia coli. J. Biol. Chem. 246, 2147–2151 (1971).
Reynolds, P. R., Mottur, G. P. & Bradbeer, C. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of btuc and tonb. J. Biol. Chem. 255, 4313–4319 (1980).
Postle, K. & Skare, J. T. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J. Biol. Chem. 263, 11000–11007 (1988).
Evans, J. S., Levine, B. A., Trayer, I. P., Dorman, C. J. & Higgins, C. F. Sequence-imposed structural constraints in the TonB protein of Escherichia coli. FEBS Lett. 208, 211–216 (1986).
Brewer, S. et al. Structure and function of X-Pro dipeptide repeats in the TonB proteins of Salmonella typhimurium and Escherichia coli. J. Mol. Biol. 216, 883–895 (1990).
Larsen, R. A., Wood, C. & Postle, K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10, 943–953 (1993).
Holroyd, C. D. & Bradbeer, C. in Microbiology (ed. Schlessinger, D.) 21–23 (American Society for Microbiology, Washington D. C., 1984).
Larsen, R. A., Thomas, M. G. & Postle, K. Protonmotive force, ExbB and ligand-FepA drive conformational changes in TonB. Mol. Microbiol. 31, 1809–1824 (1999). This paper presents evidence that supports a mechanism whereby TonB cycles between different conformations in response to the proton gradient across the cytoplasmic membrane, and proposes a model of energy transduction to the outer-membrane receptors.
Skare, J. T., Ahmer, B. M. M., Seachord, C. L., Darveau, R. P. & Postle, K. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268, 16302–16308 (1993).
Larsen, R. A. et al. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J. Bacteriol. 178, 1363–1373 (1996).
Cadieux, N. & Kadner, R. J. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl Acad. Sci. USA 96, 10673–10678 (1999).
Higgs, P. I. et al. TonB interacts with non-receptor proteins at the outer membrane of Escherichia coli. J. Bacteriol. 184, 1640–1648 (2002).
Kampfenkel, K. & Braun, V. Membrane topology of the Escherichia coli ExbD protein. J. Bacteriol. 174, 5485–5487 (1992).
Kampfenkel, K. & Braun, V. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 268, 6050–6057 (1993).
Higgs, P. I., Myers, P. S. & Postle, K. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J. Bacteriol. 180, 6031–6038 (1998).
Higgs, P. I., Larsen, R. A. & Postle, K. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44, 271–281 (2002).
Held, K. G. & Postle, K. ExbB and ExbD do not function independently in TonB-dependent energy transduction. J. Bacteriol. 184, 5170–5173 (2002). References 77 – 79 are concerned with the topological characterization of the energy-transducing complex that is formed by the proteins TonB, ExbB and ExbD.
Karlsson, M., Hannavy, K. & Higgins, C. F. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8, 389–396 (1993).
Traub, I., Gaisser, S. & Braun, V. Activity domains of the TonB protein. Mol. Microbiol. 8, 409–423 (1993).
Larsen, R. A., Thomas, M. G., Wood, G. E. & Postle, K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol. Microbiol. 13, 627–640 (1994).
Braun, V. et al. Energy coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178, 2836–2845 (1996).
Larsen, R. A. & Postle, K. Conserved residues Ser16 and His20 and their relative positioning are essential for TonB activity, cross-linking of TonB with ExbB and the ability of TonB to respond to proton motive force. J. Biol. Chem. 276, 8111–8117 (2001).
Chang, C., Mooser, A., Plückthun, A. & Wlodawer, A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J. Biol. Chem. 276, 27535–27540 (2001). This paper reports the atomic structure of a carboxy-terminal fragment of TonB, revealing an unexpected dimeric form.
Moeck, G. & Letellier, L. Characterization of in vitro interactions between a truncated TonB protein from Escherichia coli and the outer membrane receptors FhuA and FepA. J. Bacteriol. 183, 2755–2764 (2001).
Neilands, J. B. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270, 26723–26726 (1995).
Pattus, F. & Abdallah, M. A. Siderophores and iron-transport in microorganisms. J. Chin. Chem. Soc. 47, 1–20 (2000).
Roosenberg, J. M., Lin, Y. M., Lu, Y. & Miller, M. J. Studies and synthesis of siderophores, microbial iron chelators and analogs as potential drug delivery agents. Curr. Med. Chem. 7, 159–197 (2000).
Ferguson, A. D. et al. Crystal structure of the antibiotic albomycin in complex with the outer membrane transporter FhuA. Protein Sci. 9, 956–963 (2000).
Ferguson, A. D. et al. Active transport of an antibiotic ryfamycin derivative by the outer membrane protein FhuA. Structure 9, 707–716 (2001).
Braun, V. & Braun, M. Active transport of iron and siderophore antibiotics. Curr. Opin. Microbiol 5, 194–201 (2002).
Clarke, T. E., Braun, V., Winkelmann, G., Tari, L. W. & Vogel, H. J. X-ray crystallographic structures of the Escherichia coli periplasmic binding protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J. Biol. Chem. 277, 13966–13972 (2002).
Kadner, R. J. in Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt, F.) 58–87 (American Society for Microbiology, Washington DC, 1996).
Oliver, D. B. in Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt, F.) 88–103 (American Society for Microbiology, Washington DC, 1996).
Park, J. T. in Escherichia coli and Salmonella typhimurium: cellular and molecular biology (ed. Neidhardt, F.) 48–57 (American Society for Microbiology, Washington DC, 1996).
Köster, W. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12 . Res. Microbiol. 152, 291–301 (2001).
Sprencel, C. et al. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182, 5359–5364 (2000).
Cadieux, N. et al. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184, 706–717 (2002).
Locher, K. P., Lee, A. T. & Rees, D. C. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1098 (2002).
Acknowledgements
This work was supported by grants by the EPSRC, The British Council and La Caixa Foundation. Our thanks to L. Forrest, F. Pattus and I. Schalk for helpful duscussions.
Author information
Authors and Affiliations
Corresponding author
Glossary
- IRON–SULPHUR PROTEIN
-
A protein that contains one or more clusters of Fe and S atoms, which have an essential role in a wide range of reduction reactions in biological systems, such as oxidative phosphorylation and photosynthesis.
- SUPEROXIDE DISMUTASE
-
An enzyme that is present in all aerobic organisms. It catalyses the conversion of the highly reactive and destructive superoxide anion radicals, which are generated by the metabolism of the cell, into hydrogen peroxide.
- TRANSFERRIN
-
An iron-binding protein that is commonly found in the physiological fluids (serum, milk, saliva, and so on) of many vertebrates. Transferrin acts as an iron carrier with potent antibacterial properties.
- HAEMOPHORE
-
A relatively small protein that is used by Gram-negative bacteria to capture iron-containing haem from complexes such as haemopexin or haemoglobin, and to shuttle it to TonB-dependent outer-membrane receptors, which mediate further uptake into the periplasmic space.
- SIDEROPHORE
-
A low-molecular-weight compound that is produced by bacteria and other microorganisms to sequester iron from other iron-containing molecules in the medium, for example transferrin in a human host.
- PORIN
-
A channel-forming β-barrel protein that resides in the outer membrane of Gram-negative bacteria and mitochondria, across which small molecules and ions diffuse, driven by electrochemical gradients.
- PROTON-MOTIVE FORCE
-
(PMF). The effective force resulting from the relaxation of gradients in the concentration of hydrogen ions across biological membranes, which typically drives functionally important conformational changes in proteins.
- GATING
-
The opening and closing of some pore-forming membrane proteins, to regulate the passage of substances across the cell membrane.
- ABC TRANSPORTER
-
An ATP-driven membrane pump found in all known organisms, the function of which is to mediate the energy-dependent translocation of substrates ranging from inorganic ions and amino acids, to complex polysaccharides and even proteins.
- ELECTRON PARAMAGNETIC RESONANCE
-
(EPR). A spectroscopic technique that, in combination with site-directed spin labelling (or substitution of amino-acid side chains by a nitroxide group), allows the study of the structural and dynamic properties of proteins, typically by providing information on the accessibility, mobility and relative distances of the spin labels used.
- CIRCULAR DICHROISM
-
(CD). The difference in absorption of left and right circularly polarized light — the shape and magnitude of the CD curve as a function of the wavelength of protein/macromolecule solutions is sensitive to changes in the conformation of these solutes.
- CHROMOPHORE
-
The part of a molecule that is responsible for light absorption over a given range of wavelengths.
- FLUORESCENCE RESONANCE ENERGY TRANSFER
-
(FRET). A spectroscopic technique that allows the study of conformational changes in proteins and protein–ligand complexes by monitoring changes in the relative distances between fluorescent groups, such as tryptophan side chains or extrinsic fluorescent probes.
- FENTON REACTION
-
A chemical reaction that occurs when transition metals such as iron interact with hydrogen peroxide. The reaction produces highly reactive and potentially damaging hydroxyl radicals.
- PERIPLASMIC-BINDING PROTEIN
-
A carrier protein found in the periplasmic space of Gram-negative bacteria and mitochondria, the function of which is to facilitate the translocation of nutrients and other compounds across the periplasm and the peptidoglycan mesh.
- SIGMA FACTOR
-
The subunit of the prokaryotic RNA polymerase that is responsible for the recognition of specific initiation sequences (promoters), which leads to gene transcription.
- SPHEROPLAST
-
A bacterial or plant cell from which most of the cell wall has been removed, usually by enzymatic treatment, but which has not lysed.
- PROTONOPHORE
-
(ionophore). A small hydrophobic compound that associates with inorganic ions and protons, and that is able to diffuse across lipid membranes, thereby reducing or abolishing electrochemical gradients across the membrane.
- AMPHIPATHIC
-
In the context of proteins, a segment that contains both hydrophobic (for example, phenylalanine) and hydrophilic (for example, arginine) amino acids.
Rights and permissions
About this article
Cite this article
Faraldo-Gómez, J., Sansom, M. Acquisition of siderophores in Gram-negative bacteria. Nat Rev Mol Cell Biol 4, 105–116 (2003). https://doi.org/10.1038/nrm1015
Issue Date:
DOI: https://doi.org/10.1038/nrm1015
This article is cited by
-
Tackling the outer membrane: facilitating compound entry into Gram-negative bacterial pathogens
npj Antimicrobials and Resistance (2023)
-
Radiochemical Evidence for the Contribution of Chemotyped Siderophore Producing Bacteria Towards Plant Iron Nutrition
Current Microbiology (2021)
-
Bacterial siderophores in community and host interactions
Nature Reviews Microbiology (2020)
-
Isolation and genomic characterization of a pathogenic Providencia rettgeri strain G0519 in turtle Trachemys scripta
Antonie van Leeuwenhoek (2020)
-
Advances in the antimicrobial and therapeutic potential of siderophores
Environmental Chemistry Letters (2019)