Key Points
-
ATP synthase is a ubiquitous, highly conserved enzyme that catalyses the formation of ATP from ADP and Pi using a unique rotary motor mechanism.
-
The enzyme is located in the inner membrane of mitochondria, in the thylakoid membrane of chloroplasts, and in the plasma membrane of bacteria.
-
Recent analysis of the crystal structure of the enzyme has shown in atomic detail the intricate mechanisms of rotary catalysis.
-
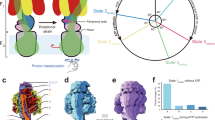
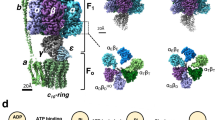
ATP synthase is a large (500 kDa) multisubunit protein, consisting of an intrinsic membrane domain, Fo, linked through central and side stalks to a globular catalytic domain, F1.
-
The F1 portion consists of three α- and three β-subunits and a single γδɛ-subunit, whereas Fo comprises one a-subunit, two b-subunits and 10–12 c-subunits.
-
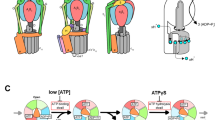
The synthesis of ATP is brought about by the rotary motion of the FoF1 complex: when a large electrochemical potential (proton gradient) flows through the Fo subunit, this causes rotation of the Fo subunit and, subsequently, F1, leading to ATP synthesis.
-
ATP hydrolysis by ATPase — the reverse reaction — induces rotation of the Fo rotor in the opposite direction. So, ATP synthase can be viewed as a complex of two motors: an ATP-driven F1 motor and the proton-driven Fo motor.
Abstract
ATP synthase can be thought of as a complex of two motors — the ATP-driven F1 motor and the proton-driven Fo motor — that rotate in opposite directions. The mechanisms by which rotation and catalysis are coupled in the working enzyme are now being unravelled on a molecular scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Boyer, P. D. The ATP synthase — a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997).Boyer's rotational catalysis and alternate-binding change model are concisely reviewed.
Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type mechanism. Nature 191, 144–148 (1961).This paper introduced a new concept of chemiosmotic theory into the field of bioenergetics.
Kanazawa, H., Kayano, T., Mabuchi, K. & Futai, M. Nucleotide sequence of the genes coding for α-, β- and γ-subunits of the proton-translocating ATPase of Escherichia coli. Biochem. Biophys. Res. Commun. 103, 604–612 (1981).
Walker, J. E., Fearnley, I. M., Gay, N. J., Gibson, B. W. & Tybulewicz, V. L. J. Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J. Mol. Biol. 184, 677–701 (1985).
Hudson, G. S. et al. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and three CFo subunits of the H+-ATP synthase complex and the ribosomal protein S2. J. Mol. Biol. 196, 283–298 (1987).
Abrahams, J. P., Leslie, A. G., Lutter, R. & Walker, J. E. Structure at 2. 8 Å of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).The demonstration of the molecular structure of the major part of the enzyme strongly indicated the rotation of the central γ-subunit surrounded by the cylinder of α 3 β 3 -subunits.
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. J. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).The striking direct demonstration of the rotation of the γ-subunit.
Tsunoda, S. P. et al. Observations of rotation within the FoF1-ATP synthase: deciding between rotation of the Foc subunit ring and artifact. FEBS Lett. 470, 244–248 (2000).
Menz, R. I., Walker, J. E. & Leslie, A. G. W. Crystal structure of bovine mitochondrial F1-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell 106, 331-341 (2001).Three bound adenine nucleotides at the catalytic sites and the slightly twisted γ-subunit led to the proposal of an intermediate structure during catalysis. | Contents Page |
Yasuda, R., Noji, H., Kinosita, K. J. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell 93, 1117–1124 (1998).
Soong, R. K. et al. Powering an inorganic nanodevice with a biomolecular motor. Science 290, 1555–1558 (2000).
Oster, G. & Wang, H. Why is the efficiency of the F1 ATPase so high? J. Bioenerg. Biomembr. 32, 459–469 (2000).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. J. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).A 120° step rotation is further divided into 90° and 30° substeps. ATP binding triggers the former and release of ADP·P i does the latter substep.
Yagi, H. et al. Functional conformation changes in the F1-ATPase β subunit probed by 12 tyrosine residues. Biophys. J. 77, 2175–2183 (1999).
Tsunoda, S. P., Muneyuki, E., Amano, T., Yoshida, M. & Noji, H. Cross-linking of two β subunits in the closed conformation in F1-ATPase. J. Biol. Chem. 274, 5701–5706 (1999).
Ren, H., Dou, C., Stelzer, M. S. & Allison, W. S. Oxidation of the α3(βD311C/R333C)3γ subcomplex of the thermophilic Bacillus PS3 F1-ATPase indicates that only two β-subunits can exist in the closed conformation simultaneously. J. Biol. Chem. 274, 31366–31372 (1999).
Kayalar, C., Rosing, J. A. N. & Boyer, P. D. An alternating site sequence for oxidative phosphorylation suggested by measurement of substrate binding patterns and exchange reaction inhibitions. J. Biol. Chem. 252, 2486–2491 (1977).
Gresser, M. J., Myers, J. A. & Boyer, P. D. Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model. J. Biol. Chem. 257, 12030–12038 (1982).
Weber, J., Wilke-Mounts, S., Lee, R. S. F., Grell, E. & Senior, A. E. Specific placement of tryptophan in the catalytic sites of Escherichia coli F1-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. J. Biol. Chem. 268, 20126–20133 (1993).
Ren, H. & Allison, W. S. On what makes the γ-subunit spin during ATP hydrolysis by F1 . Biochim. Biophys. Acta 1458, 221–233 (2000).
Hackney, D. D., Rosen, G. & Boyer, P. D. Subunit interaction during catalysis: alternating site cooperativity in photophosphorylation shown by substrate modulation of [18O]ATP species formation. Proc. Natl Acad. Sci. USA 76, 3646–3650 (1979).
Hackney, D. D. & Boyer, P. D. Subunit interaction during catalysis. Implications of concentration dependency of oxygen exchanges accompanying oxidative phosphorylation for alternating site cooperativity. J. Biol. Chem. 253, 3164–3170 (1978).
Zhou, Y., Duncan, T. M. & Cross, R. L. Subunit rotation in Escherichia coli FoF1-ATP synthase during oxidative phosphorylation. Proc. Natl Acad. Sci. USA 94, 10583–10587 (1997).
Stock, D., Leslie, A. G. W. & Walker, J. E. Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 (1999).
Sambongi, Y. et al. Mechanical rotation of the c subunit oligomer in ATP synthase (FoF1): direct observation. Science 286, 1722–1724 (1999).
Pänke, O., Gumbiowski, K., Junge, W. & Engelbrecht, S. F-ATPase: specific observation of the rotating c subunit oligomer of EFoEF1 . FEBS Lett. 472, 34–38 (2000).
Tsunoda, S. P., Aggeler, R., Yoshida, M. & Capaldi, R. A. Rotation of the c subunit oligomer in fully functional F1Fo ATP synthase. Proc. Natl Acad. Sci. USA 98, 898–902 (2001).
Hutcheon, M. L., Duncan, T. M., Ngai, H. & Cross, R. L. Energy-driven subunit rotation at the interface between subunit a and the c oligomer in the Fo sector of Escherichia coli ATP synthase. Proc. Natl Acad. Sci. USA 98, 8519–8524 (2001).
Rastogi, V. K. & Girvin, M. K. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature 402, 263–268 (1999).
Seelert, H. et al. Structural biology. Proton-powered turbine of a plant motor. Nature 405, 418–419 (2000).
Stahlberg, H. et al. Bacterial Na+-ATP synthase has an undecameric rotor. EMBO Rep. 2, 229–233 (2001).
Jiang, W., Hermolin, J. & Fillingame, R. H. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl Acad. Sci. USA 98, 4966–4971 (2001).
Schemidt, R. A., Qu, J., Williams, J. R. & Brusilow, W. S. Effects of carbon source on expression of Fo genes and on the stoichiometry of the c subunit in the F1Fo ATPase of Escherichia coli. J. Bacteriol. 180, 3205–3208 (1998).
Sorgen, P. L., Bubb, M. R. & Cain, B. D. Lengthening the second stalk of F1Fo ATP synthase in Escherichia coli. J. Biol. Chem. 274, 36261–36266 (1999).
Elston, T., Wang, H. & Oster, G. Energy transduction in ATP synthase. Nature 391, 510–513 (1998).
Dimroth, P., Wang, H., Grabe, M. & Oster, G. Energy transduction in the sodium F-ATPase of Propionigenium modestum. Proc. Natl Acad. Sci. USA 96, 4924–4929 (1999).
Ruppert, C. et al. The proteolipid of the A1A0 ATP synthase from Methanococcus jannaschii has six predicted transmembrane helices but only two proton-translocating carboxyl groups. J. Biol. Chem. 274, 25281–25284 (1999).
Aufurth, S., Schagger, H. & Muller, V. Identification of subunits a, b, and c1 from Acetobacterium woodii Na+-F1Fo-ATPase. Subunits c1, c2, and c3 constitute a mixed c-oligomer. J. Biol. Chem. 275, 33297–33301 (2000).
Junge, W., Lill, H. & Engelbrecht, S. ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 (1997).
Miller, M. J., Oldenburg, M. & Fillingame, R. H. The essential carboxyl group in subunit c of F1Fo ATP synthase can be moved and H+-translocating function retained. Proc. Natl Acad. Sci. USA 87, 4900–4904 (1990).
Nalin, C. M. & McCarty, R. E. Role of a disulfide bond in the γ-subunit in activation of the ATPase of chloroplast coupling factor 1. J. Biol. Chem. 259, 7275–7280 (1984).
Werener-Gruene, S., Gunkel, D., Schumann, J. & Strotmann, H. Insertion of a chloroplast-like regulatory segment responsible for thiol modulation into γ-subunit of FoF1-ATPase of the cyanobacterium Synechocystis 6803 by mutagenesis of atpC. Mol. Gen. Genet. 244, 144–150 (1994).
Bald, D., Noji, H., Stumpp, M. T., Yoshida, M. & Hisabori, T. ATPase activity of a highly stable α3β3γ subcomplex of thermophilic F1 can be regulated by the introduced regulatory region of γ-subunit of chloroplast F1 . J. Biol. Chem. 275, 12757–12762 (2000).
Lebowitz, M. S. & Pedersen, P. L. Protein inhibitor of mitochondrial ATP synthase: relationship of inhibitor structure to pH-dependent regulation. Arch. Biochem. Biophys. 330, 342–354 (1996).
Cabezon, E., Arechaga, I., Jonathan, P., Butler, G. & Walker, J. E. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1 . J. Biol. Chem. 275, 28353–28355 (2000).
Cabezon, E., Butler, P. J., Runswick, M. J. & Walker, J. E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 275, 25460–25464 (2000).
Jault, J. M. & Allison, W. S. Slow binding of ATP to noncatalytic nucleotide binding sites which accelerate catalysis is responsible for apparent negative cooperativity exhibited by the bovine mitochondrial F1-ATPase. J. Biol. Chem. 268, 1558–1566 (1993).
Matsui, T. et al. Catalytic activity of the α3β3γ complex of F1-ATPase without noncatalytic nucleotide binding site. J. Biol. Chem. 272, 8215–8221 (1997).
Minkov, I. B., Vasilyeva, E. A., Fitin, A. F. & Vinogradov, A. D. Differential effects of ADP on ATPase and oxidative phophorylation in submitochondrial particles. Biochem. Int. 1, 478–485 (1980).
Bald, D. et al. ATP synthesis by FoF1-ATP synthase independent of noncatalytic nucleotide binding sites and insensitive to azide inhibition. J. Biol. Chem. 273, 865–870 (1998).
Rodgers, A. J. & Wilce, M. C. Structure of the γ–ɛ complex of ATP synthase. Nature Struct. Biol. 7, 1051–1054 (2000).
Gibbons, C., Montgomery, M. G., Leslie, A. G. & Walker, J. E. The structure of the central stalk in bovine F1-ATPase at 2.4-Å resolution. Nature Struct. Biol. 7, 1055–1061 (2000).
Wilkens, S. & Capaldi, R. A. Solution structure of the ɛ-subunit of the F1-ATPase from Escherichia coli and interactions of this subunit with β-subunits in the complex. J. Biol. Chem. 273, 26645–26651 (1998).
Hara, K. Y., Kato-Yamada, Y., Kikuchi, Y., Hisabori, T. & Yoshida, M. The role of the βDELSEED motif of F1–ATPase; propagation of the inhibitory effect of the ɛ-subunit. J. Biol. Chem. 276, 23969–23973 (2001). |
Kato-Yamada, Y., Yoshida, M. & Hisabori, T. Movement of the helical domain of the ɛ subunit is required for the activation of thermophilic F1-ATPase. J. Biol. Chem. 275, 35746–35750 (2000).
Tsunoda, S. P. et al. Large conformational changes of the ɛ subunit in the bacterial F1Fo ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl Acad. Sci. USA 98, 6560–6564 (2001).
Lohse, D. & Strotmann, H. Reaction related with ΔpH-dependent activation of the chloroplast H+-ATPase. Biochim. Biophys. Acta 976, 94–101 (1989).
Galkin, M. A. & Vinogradov, A. D. Energy-dependent transformation of the catalytic activities of the mitochondrial Fo × F1-ATP synthase. FEBS Lett. 448, 123–126 (1999).
Fischer, S., Gräber, P. & Turina, P. The activity of the ATP synthase from Escherichia coli is regulated by the transmembrane proton motive force. J. Biol. Chem. 275, 30157–30162 (2000).
Kaim, K. & Dimroth, P. ATP synthesis by F-type ATP synthase is obligatorily dependent on the transmembrane voltage. EMBO J. 18, 4118–4127 (1999).
Pullman, M. E., Penefsky, H. S., Datta, A. & Racker, E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble, dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 235, 3322–3329 (1960).
Penefsky, H. S., Pullman, M. E., Datta, A. & Racker, E. Partial resolution of the enzyme catalyzing oxidative phosphorylation. II. Participation of a soluble adenosine triphosphatase in oxidative phosphorylation. J. Biol. Chem. 235, 3330–3336 (1960).
Mitchell, P. Keilin's respiratory chain concept and its chemiosmotic consequences. Science 206, 1148–1159 (1979). |
Jagendorf, A. T. & Uribe, E. ATP formation caused by acid-base transition of spinach chloroplasts. Proc. Natl Acad. Sci. USA 55, 170–177 (1966).The turning point for the Mitchell's chemiosmotic theory. After this, many people began to regard the chemiosmotic theory as the strongest hypothesis for oxidative and photo-phosphorylation.
Kagawa, Y. & Racker, E. Partial resolution of the enzyme catalyzing oxidative phosphorykation. XXV. Reconstitution of vesicles catalyzing 32Pi- adenosine triphosphate exchange. J. Biol. Chem. 246, 5477–5487 (1971).The most convincing evidence for the chemiosmotic theory. The reconstitution method of membrane proteins described here had a profound influence over the field of membrane biochemistry.
Sone, N., Yoshida, M., Hirata, H. & Kagawa, Y. Adenosine triphosphate synthesis by electrochemical proton gradient in vesicles reconstituted from purified adenosine triphosphatase and phospholipids of thermophilic bacterium. J. Biol. Chem. 252, 2956–2960 (1977).
Boyer, P. D. Energy, life and ATP. Angew. Chem. Int. Ed. 37, 2296–2307 (1998).
Grubmeyer, C., Cross, R. L. & Penefsky, H. S. Mechanism of ATP hydrolysis by beef heart mitochondrial ATPase. Rate constants for elementary steps in catalysis at a single site. J. Biol. Chem. 257, 12092–12100 (1982).
Walker, J. E. ATP Synthesis by Rotary Catalysis, 208–234 (The Nobel Foundation, Stockholm, 1997).
Duncan, T. M., Bulygin, V. V., Zhou, Y., Hutcheon, M. L. & Cross, R. L. Rotation of subunits during catalysis by Escherichia coli F1-ATPase. Proc. Natl Acad. Sci. USA 92, 10964–10968 (1995).
Iwata, S. et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71 (1998).
Tsukihara, T. et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269, 1069–1074 (1995).
Jagendorf, A. T. & Smith, M. Uncoupling phosphorylation in spinach chloroplasts by absence of cations. Plant Physiol. 37, 135–141 (1962).
Fessenden, J. M. & Racker, E. Partial resolution of the enzyme catalyzing oxidative phosphorylation. XI. Stimulation of oxidative phosphorylation by coupling factors and oligomycin; inhibition by an antibody against coupling factor 1. J. Biol. Chem. 241, 2483–2489 (1966).
Adachi, K. et al. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl Acad. Sci. USA 97, 7243–7247 (2000).
Uhlin, U., Cox, G. B. & Guss, J. M. Crystal structure of the ɛ subunit of the proton-translocating ATP synthase from Escherichia coli. Structure 5, 1219–1230 (1997).
Acknowledgements
We are grateful to A. Leslie and J. Walker for providing us with the preprint of their paper on the structure of (ADP·AlF4−)2F1. We also thank T. Suzuki and K. Tsukuda for their assistance in the preparation of the figures.
Author information
Authors and Affiliations
Related links
Glossary
- ELECTROCHEMICAL POTENTIAL GRADIENT
-
When two aqueous phases are separated by a membrane, the electrochemical potential difference of H+ between the two phases is expressed as Δ\(\overline{μ}\)H+ = FΔΨ−2.3RTΔpH, where F is the Faraday constant, ΔΨ is the electric potential difference between two phases, R is the gas constant, T is the absolute temperature and ΔpH is pH difference between two phases.
- SWITCH II REGION
-
The β-subunit of F1 has a region that is topologically equivalent to the switch II region of guanine-nucleotide binding (G) proteins, which changes the conformation in response to the interconversion of GTP and GDP.
- P-LOOP
-
Various ATP-metabolizing proteins contain a consensus sequence Gly-X-X-Gly-X-Gly-Lys-Thr (X is variable). This sequence is found in a loop connecting a β-strand (adjacent to a β-strand of switch II region) and an α-helix. The lysine and threonine residues in the P-loop are recruited for binding the phosphate moiety of nucleotides.
- V-ATPASE
-
V-ATPase is responsible for ATP synthesis in archaebacteria and a small number of eubacteria. In eukaryotic cells, it works as a proton-translocating machinery driven by ATP hydrolysis, and it is responsible for the acidification of lysosome lumens, chromaffin granules and vacuoles.
Rights and permissions
About this article
Cite this article
Yoshida, M., Muneyuki, E. & Hisabori, T. ATP synthase — a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol 2, 669–677 (2001). https://doi.org/10.1038/35089509
Issue Date:
DOI: https://doi.org/10.1038/35089509
This article is cited by
-
Multi-omics analysis identifies drivers of protein phosphorylation
Genome Biology (2023)
-
Molecular mechanism on forcible ejection of ATPase inhibitory factor 1 from mitochondrial ATP synthase
Nature Communications (2023)
-
Sustained unidirectional rotation of a self-organized DNA rotor on a nanopore
Nature Physics (2022)
-
The V-ATPases in cancer and cell death
Cancer Gene Therapy (2022)
-
RNA-Seq Transcriptomic Analysis of Green Tea Polyphenols Modulation of Differently Expressed Genes in Enterococcus faecalis Under Bile Salt Stress
Current Microbiology (2022)