Abstract
One of the main challenges of developing an HIV-1 vaccine lies in eliciting immune responses that can overcome the antigenic variability exhibited by HIV. Most HIV-1 vaccine development has focused on inducing immunity to conserved regions of the HIV-1 envelope. However, new studies of the sequence-variable regions of the HIV-1 gp120 envelope glycoprotein have shown that there are conserved immunological and structural features in these regions. Recombinant immunogens that include these features may provide the means to address the antigenic diversity of HIV-1 and induce protective antibodies that can prevent infection with HIV-1.
Similar content being viewed by others
Main
The gp120 and gp41 glycoproteins of the HIV envelope (Env) are targets of antibodies that inhibit virus infectivity, and attempts to induce these antibodies with a prophylactic HIV vaccine have used various approaches. The failure of early gp120 protein-based vaccines to induce neutralizing antibodies1 redirected attention to vaccine-mediated induction of the cellular arm of the immune response. The subsequent failure of a T cell-based vaccine2 left the HIV-1 vaccine field at a crossroads, with uncertainty as to the way forward. Recently, a dual-component HIV-1 vaccine, in which priming with a recombinant canarypox vector was followed by boosts with two recombinant gp120 proteins, imparted a measure of protection from infection3. Protection in this trial may have been due to the induction of an antibody response4, but the nature and specificity of the protective antibodies, and how to design immunogens that induce higher and more persistent levels of such antibodies, remain unresolved issues.
The difficulties of HIV-1 vaccine research are, in part, a result of the extreme antigenic variation of HIV-1. Conventional wisdom suggests that 'constant' rather than 'variable' regions of Env should be targeted by the immune system to elicit broad responses against antigenically diverse HIV strains. However, these regions were classified in early studies on the basis of the sequences of only a few virus strains5. Immunological and three-dimensional (3D) structural studies of Env from diverse strains of HIV-1 have now revealed the presence of higher-order structural features that would alter these early classifications and explain how antibodies specific for some variable regions have neutralizing activity against diverse viruses. Although several epitopes that induce neutralizing antibodies have been identified in conserved regions of both the gp120 and gp41 portions of Env, inducing neutralizing antibodies that target these epitopes using rationally designed immunogens has, so far, been difficult6,7,8,9. Moreover, although a vigorous antibody response is induced by natural infection with HIV-1, only a minority of individual HIV-positive patient sera neutralize a broad range of HIV strains10,11.
We think that targeting conserved elements within the immunogenic sequence-variable Env regions (Fig. 1) using rationally designed immunogens is a promising approach for vaccine design, but inducing broad, cross-strain neutralizing antibodies specific for these regions is a challenge. Nevertheless, current evidence indicates that 3D visualization might identify additional invariant epitopes that are capable of inducing protective antibodies and that are hidden within the sequence-variable Env regions. Indeed, this view provides a rational basis for understanding the documented immunological cross-reactivity of many monoclonal antibodies targeting the second (V2) and third (V3) variable loops of gp120, and the quaternary neutralization epitopes (QNEs) formed by V2 and V3 (Box 1; Table 1). In hindsight, this perspective is a logical outgrowth of classic immunochemical studies12 showing that cross-reactive antibodies recognize antigens that are related but not identical in sequence (as is the case for V2-specific monoclonal antibodies that recognize gp120 monomers derived from diverse primary isolates of HIV-1 (Ref. 13)). These old, but seminal, studies support a new paradigm for HIV-1 immunogen design.
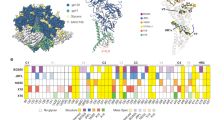
a | This gp120 monomer structure represents the truncated, deglycosylated conserved core of gp120 without the variable loops. The locations of the variable region stems and bridging sheet are shown. b | Ribbon diagram of the crystallographic structure of the V3 loop of gp120 in situ from the crystal structure, indicating the base, stem and crown regions of the V3 loop. c | Zones of sequence variability mapped onto the β-hairpin conformation of the V3 crown. The crystallographic structure of the V3 crown is shown as a ribbon diagram, with commonly occurring side chains shown in stick depiction. A multicoloured illustrative cylinder is superimposed on the structure to highlight the four zones of the V3 crown: the arch (green), the hydrophilic face (red) and hydrophobic face (blue) of the circlet, and the band (yellow). Only the hydrophilic face has high sequence variability, but the amino acids that constitute the region of variability are distributed widely in the linear protein sequence, which obscures the existence of such a region unless the 3D structural context is appreciated. The side chains of the V3 crown that are bound by neutralizing monoclonal antibodies are annotated with coloured lines: monoclonal antibody 268 neutralizes only a few HIV-1 strains and engages various side chains, including one in the variable hydrophilic face of V3; monoclonal antibodies 2219 and 3074 neutralize viruses from several HIV-1 subtypes37 and avoid engagement of the hydrophilic variable zone. The image in part a was drawn using data from Ref. 17. The image in part b was drawn using data from Ref. 80. The image in part c is adapted, with permission, from Ref. 40 © (2010) Mary Ann Liebert, Inc.
In this article, we bring together data from both the biological arena (viral and immunological studies) and the physical arena (structural and bioinformatics studies) to show how structure–function relationships of Env proteins can inform the design of an effective HIV-1 vaccine. With special emphasis on the variable regions of gp120, we highlight the relationship between 3D molecular structure, biological function and immunological cross-reactivity. This approach shows the potential utility of targeting sequence-variable regions and QNEs that are formed by the interaction of sequence-variable regions in the context of the Env trimers. Because of their sequence variability, these regions were previously dismissed as vaccine targets, but we now think that their potential use should be reassessed.
Biological functions of the gp120 regions
Antibodies that interfere with key biological functions of HIV-1 can 'neutralize' virus infectivity. Therefore, understanding the function of various parts of Env assists in determining the antibody targets that should be induced by protective vaccines.
Receptor-binding sites. It is now well established that to trigger exposure of the HIV-1 gp41 fusion domain that initiates virus–host cell fusion, gp120 must first bind to two cell-surface receptors: CD4 and CXC-chemokine receptor 4 (CXCR4) or CC-chemokine receptor 5 (CCR5)14,15. The receptor-binding surfaces on gp120 form distinct structural and antigenic regions that are highly conformational and are formed by discontinuous amino acids in several regions of gp120 (Refs 16, 17 (Box 1). The CD4-binding site consists of many residues in the constant regions of gp120 but, until recently, only one of many CD4-binding site-specific monoclonal antibodies had been described with broad neutralizing activity18. Three more have recently been selected and characterized19,20(Table 1). This suggests that the crucial region within the large CD4-binding site may be difficult to target with vaccine constructs, a fact that is supported by the disappointing results from attempts to induce neutralizing CD4-binding site-specific antibodies with various vaccine constructs6,7,9.
The chemokine receptor-binding site of gp120 consists of the invariant β2 and β3 strands in the V1–V2 double loop, the β20 and β21 strands in the fourth conserved region (C4), and residues in the V3 loop16,17. This highly conformational, discontinuous region of gp120 is often known as the CD4-induced region because, as its name implies, it is present after gp120 has bound CD4 with high affinity. Many human monoclonal antibodies specific for the CD4-induced region have been described21,22,23,24, indicating the strong immunogenicity of this region. However, when exposed on the virus particle, the CD4-induced region is sterically protected from most specific antibodies during the process of infection owing presumably to its close proximity to the cell membrane25. Consequently, antibodies specific for the CD4-induced region may not protect against infection by most HIV-1 strains.
The V3 loop. The V3 loop of gp120 is a component of the chemokine receptor-binding region, and it is an important determinant of co-receptor usage26,27,28,29,30,31. The length of the V3 loop is essentially constant at 34–35 amino acids. Although its amino acid sequence is highly variable — except in clade C viruses where the V3 sequence is generally conserved32 — sequence variation is restricted to only ∼20% of the amino acid positions and is located mainly in the two β-strands in the crown of the V3 loop (Figs 1b, 2d). The functional importance of V3 is shown by the fact that deletion of V3 completely abrogates virus infectivity33. Nonetheless, the efficacy of V3-specific antibodies in blocking virus infection has been controversial because some V3-specific antibodies, particularly those elicited early after infection, have limited breadth of neutralization in terms of different strains of HIV-1 (Ref. 34): many V3-specific antibodies neutralize only the most neutralization-sensitive 'Tier 1' viruses19,35, no single V3-specific monoclonal antibody neutralizes more than ∼10–25% of the more resistant 'Tier 2' viruses19,36,37 and the accessibility of V3 is poor on many viruses38,39. However, given the essential function of V3 in HIV infectivity, its structural conservation40,41, its strong immunogenicity42, the existence of many V3-specific monoclonal antibodies with cross-clade neutralizing activity19,36,37,43 and the demonstration that polyclonal V3-specific antibodies can be induced that neutralize viruses from diverse HIV-1 subtypes44, the V3 region remains a potentially important target for vaccine development.
a | Sequence logo depicting the amino acid conservation pattern across a multiple alignment of many V1 loops, each of which was selected because it has the most common V1 length depicted in Fig. 3a. Data used to derive the sequence logo were obtained from the Los Alamos National Laboratory (LANL), New Mexico, USA, using one sequence per patient, and all HIV-1 subtypes were included. The height of the letter indicates the degree of conservation of the most common amino acid at that position. Amino acids are coloured according to their chemical properties: small amino acids (G, P, A, S and T) are green; strongly polar, charged basic or charged acidic amino acids (K, R, D, E, Q and N) are black and hydrophobic amino acids (V, L, I, Y, W, F and M) are blue. The few amino- and carboxy-terminal amino acids of the V1 loop are reasonably conserved, but most of the loop fades to small letters, which indicate no conservation except for a run of asparagine (N) and threonine (T) residues near to the centre of the loop, suggesting a region of putative glycosylation. b | Sequence logo depicting the amino acid conservation pattern across a multiple alignment of many V2 loops, each of which was selected because it has the most common V2 length (as depicted in Fig. 3b). Sequence data were obtained from LANL (one V2 sequence per patient, with sequences from all HIV-1 subtypes included). Although the V2 loop has many more insertions and deletions than the V3 loop, it has a degree of amino acid conservation similar to that of the V3 loop when one controls for loop length as explained above. c | The amino acid positions and sites of putative glycosylation that are implicated in binding of the V2-specific monoclonal antibody 697-D and the QNE-specific monoclonal antibodies PG9, PG16 and 2909 are mapped onto a schematic illustration of the V1–V2 loop structure. Square brackets indicate more than one commonly occurring amino acid at a single position. The individual sites associated with binding of each single antibody are distributed throughout the V1–V2 primary sequence linearly but, in 3D space, must cluster into one or a few overlapping epitopes. Note that the amino acid numbering in Fig. 2c does not exactly match that in Figs 2a and 2b owing to the variation in V1 and V2 lengths. d | Sequence logo produced in the same manner as Figs 2a and 2b depicting the amino acid conservation pattern across a multiple alignment of all V3 loops from LANL sequence data (one V3 sequence per patient, with sequences from all HIV-1 subtypes included).
The V2 loop. By contrast, V2 is not essential for virus infectivity33, which suggests that the evolutionary constraints to preserve an invariant V2 structure are less pronounced than for V3. However, V2 participates in several Env functions: by providing a motif that binds to α4β7 integrin (the receptor potentially involved in the homing of the virus to gut mucosal cells)45, by contributing to Env trimer formation46 and by participating as part of the chemokine receptor-binding surface. V2 also has a crucial role in masking neutralizing epitopes of gp120: changes in V2 length and glycosylation patterns allow viruses to escape from neutralization mediated by antibodies specific for V3 and the CD4-binding site of gp120 (Refs 47, 48, 49, 50). Given these functions of the V2 loop, its newly recognized role as a component of QNEs51,52,53,54 and its partial structural conservation (see below), V2 becomes an additional potential target for vaccine development.
V1, V4 and V5. The functions of the other variable regions of gp120 are poorly defined. V1 might have functional significance as it can be a neutralizing epitope, and several potent neutralizing V1-specific monoclonal antibodies have been developed in transgenic mice55. The immunogenicity of V1 is affected by glycosylation56, and mutations in V1 can affect virus-induced syncytium formation of host cells57; however, little more than this is known about the function of V1. V4, by contrast, has no well-defined function, although it is a target for early autologous neutralizing antibodies58 and V4-specific neutralizing antibodies have been described in immunized rabbits59. V4 is involved in neutralization escape60, and it undergoes extreme variation in early infection61. The V5 region, the only variable region that does not form a disulphide-linked loop, participates in forming the CD4-binding site surface, but no polyclonal or monoclonal neutralizing V5-specific antibodies have been described; nonetheless, V5 is involved in neutralization escape, although V5 peptides could not block the activity of neutralizing antibodies in sera62. Current data, therefore, do not support the use of these variable regions as targets for HIV-1-specific antibody-based vaccines.
Trimeric Env spikes. Crystal structures of gp120 monomers have been available for more than a decade17, but the crystal structure of the trimeric Env spike is not available, although it has been modelled using monomer structures and cryo-electron tomography63,64,65. There are important consequences to the oligomerization of the HIV-1 Env protein: trimer formation is a requirement for infectivity, results in the burial of neutralizing epitopes within oligomeric interfaces and allows conformational epitope masking66,67,68. The QNEs that form after trimerization of gp120 have been defined by various human and macaque monoclonal antibodies, which are exceptionally potent51,52,53,54, a fact that reflects a crucial function of the QNEs that has not yet been defined.
Variable loop conservation
Recently, several 3D structural studies have provided important insights into the conserved structural elements in the variable loops of gp120, which have indicated potential windows of opportunity for designing immunogens expressing these conserved structures.
The V3 loop. Although the V3 loop is the least variable of the HIV-1 Env variable regions because, in part, it is the only one that is essentially constant in length, its sequence varies considerably across strains, and this variability is most pronounced in its 'crown', ∼14 amino acids at the tip of the V3 loop (Fig. 1b). Although the specificity of some V3-specific antibodies restricts their immunochemical and biological functions69,70, some V3-specific monoclonal and polyclonal antibodies from infected individuals can neutralize diverse HIV-1 strains19,36,43,71,72,73,74. Furthermore, some individual V3-specific monoclonal antibodies can neutralize ∼25–50% of Tier 1 and Tier 2 HIV-1 strains in which the correct epitope is present19,37,43,72,75; polyclonal V3-specific responses to infection can neutralize Tier 1 and Tier 2 viruses from diverse clades74. Vaccine-induced immune responses targeted to V3 can also elicit broader neutralizing activity than that seen in many human HIV-positive sera or with mixtures of human monoclonal antibodies44.
Interestingly, crystallographic structures of the gp120 V3 loop crown from different virus strains in complex with various human monoclonal antibodies41,59,76,77,78,79, and structures of the V3 loop freely arranged in situ on the core of gp120 (Ref. 80), all have a V3 crown that forms variations of a common β-hairpin tertiary structure. Furthermore, the sequence variation of the V3 loop tends to cluster into a single continuous small zone when viewed in 3D space40 (Fig. 1c). We hypothesize that the crown of the V3 loop might be organized into a folded (albeit highly flexible) globular domain, with most of its surface being structurally, and therefore potentially antigenically, invariant. This structural perspective explains both the cross-reactivity of many V3-specific antibodies, which bind the conserved surfaces of V3, and the narrow specificity of some V3-specific monoclonal antibodies, which bind the small sequence-variable zone40,41 (Fig. 1c). The discovery of conserved 3D structures in the sequence-variable V3 loop is consistent with the function of the V3 loop as a chemokine receptor-binding element: the receptor-binding surface of gp120 must be conserved to preserve virus infectivity, and these constraints apparently involve more of the amino acid positions of the V3 loop than they leave free to vary. This view of the V3 loop indicates which regions of V3 should be targeted with immunogens to elicit broadly neutralizing antibodies; it also indicates the region of V3 that induces only strain-specific antibody responses.
The V1–V2 double loop. V2 varies in both sequence and length (Figs 2, 3) and, as a result, is a much more variable region than is V3. V2 is immunogenic in ∼20–45% of HIV-1-infected humans13 in contrast to V3, which induces antibodies in essentially all infected individuals42. Several V2-specific human monoclonal antibodies have been generated that cross-react with gp120 molecules from diverse HIV-1 isolates, and V2-specific monoclonal antibodies have also been produced from cells of infected chimpanzees and immunized rats81,82,83; this indicates that V2 might also contain some structurally conserved elements. Interestingly, many V2-specific monoclonal antibodies interact with the same amino acid residues or with overlapping regions of V2 (Refs 13, 52, 81, 83, 84) (Fig. 2c). Published studies document the neutralizing activity of human and chimpanzee V2-specific monoclonal antibodies; although many of these antibodies have little or weak neutralizing activity, some have notable potency83,85,86.
The histograms show the number (y axis) of recorded viruses having each V1 or V2 length plotted against the number of amino acids of the V1 or V2 loop (x axis). Loop length is plotted from zero to the maximal recorded length; so, for example, the origin shows that no viruses are recorded to have entirely deleted V1 or V2 loops (length of loop = 0).
Little is known about the 3D structure of the V2 loop, but the biological advantage conferred by the integrin α4β7-binding motif in V2 (Ref. 45) might constrain at least part of the V2 structure. This receptor-binding motif occurs in a region of V2 containing alternating and periodic conserved charged and hydrophobic amino acid residues that are typical of a folded domain. In addition, V2 contains a well-conserved disulphide bond; such bonds are common in small, loosely folded domains with high-level sequence variation in their 3D structural folds (such as the thioredoxin fold found in many proteins with known 3D structures)87. It is also notable that the carboxy-terminal portion of the stem of the V2 loop is located directly adjacent to the CD4-binding site in crystal structures of gp120 bound to CD417,80 and that some data indicate that the presence or absence of V2 might affect recognition and neutralization by the CD4-binding site-specific antibody IgG1b12 (Refs 66, 88). Further structural information is derived from a comparison of the length distributions of V1 and V2 (Fig. 3a,b). The normal distribution of V1 lengths is typical of random sampling from several optional structures and thereby suggests that V1 is likely to be a dynamic, unfolded and disordered flexible loop that alternates between many conformations. The only other structural clue available for V1 comes from the presence in the mid-region sequence of V1 (as observed in the most common length) of repetitive sites of putative glycosylation (Asn-X-Thr/Ser shown as V1 residues 136–144; see Fig. 2a).
The different patterns of length distribution of V1 and V2 reflect biological functions that suggest 3D structural order within V2 but, as noted, structural disorder within V1. Specifically, the V2 length distribution pattern has a sharp drop-off in length from the most frequent length to shorter lengths, but a more gradual drop-off as the V2 length increases, indicating a viral mechanism that has little tolerance of V2 lengths shorter than ∼40 amino acids. This pattern of length variation is typical of either a folded globular domain, which requires a crucial length below which the fold cannot assemble, or inter-monomer contacts, which require a crucial length below which the loop cannot reach over from one monomer to another. In both cases, the observed length distribution requires some V2 structural order. The available structural data — together with the observed cross-reactive immunological patterns of V2-specific antibodies — suggest that a similar, but more challenging, approach to vaccine targeting as envisioned for V3 might also be successful for V2, but the data also suggest that V1 is probably not a suitable vaccine target.
V2–V3 complexes in the envelope trimer. There is now evidence that V2 and V3 can, together, form complex epitopes comprised of contributions from both variable loops. These complex epitopes exist preferentially or exclusively in the trimeric form of the HIV Env spike, which defines them as quaternary epitopes (Box 1). Antibodies specific for quaternary epitopes were first described in simian–HIV89.6P (SHIV89.6P)-infected macaques; although these antibodies were difficult to characterize in polyclonal sera, their activity seemed to map to a discontinuous epitope formed by V2 and V3 present on the Env spikes of the virus89. Similar antibodies were found in an HIV-infected chimpanzee90,91. Remarkably, these antibodies were extremely potent neutralizing reagents. The first human QNE-specific monoclonal antibody to be described, 2909, is extremely potent (IC50 < 1.0 ng/ml) and targets both the V2 and V3 regions of gp120 (Ref. 51). The epitope to which monoclonal antibody 2909 binds is found only on the surface of virions and on env-transfected cells and does not exist on the gp120 monomer or recombinant soluble Env trimers52,68, which indicates that this epitope is formed only when gp120 monomers form trimers in lipid membranes. Monoclonal antibody 2909 is highly strain specific although it tolerates extensive amino acid sequence changes in V3 (Ref. 52). The strain specificity of 2909 is the result of binding to a lysine, rather than the more common asparagine, at residue 160 in V2 (Ref 52, 54) (Fig. 2b,c). So, although 2909 is narrow in its reactivity, this monoclonal antibody defined the first quaternary epitope in the HIV-1 Env protein and revealed both tolerance for extensive variation in V3 and specificity constraints imposed by a single position in V2.
Recently, several QNE-specific monoclonal antibodies have been generated from macaques infected with SHIVSF162P4 (Ref. 54). Similar to monoclonal antibody 2909, these antibodies have potent but strain-specific neutralizing activity, target V2 and V3, and are limited in terms of their breadth of reactivity by residues in V2. So, macaques and humans both make similar potent QNE-specific antibodies.
Notably, two clonally related human monoclonal antibodies (PG9 and PG16) have now been described that also target V2 and V3 and have potent neutralizing activity (IC50 < 1.0 ng/ml for many strains)53. Both of these monoclonal antibodies can be considered QNE-specific antibodies although PG9 reacts weakly with monomeric gp120 as well as with trimeric gp140 and seems to target mainly discontinuous residues in gp120 V2. Monoclonal antibody PG16, which is a true QNE-specific antibody, recognizes only the trimeric form of gp120 and interacts with discontinuous residues found in V2 and the crown of V3. Unlike human monoclonal antibody 2909 and the macaque QNE-specific monoclonal antibodies, PG9 and PG16 have broad neutralizing activity, neutralizing 73% and 79% of 162 diverse pseudoviruses, respectively. The crucial difference that distinguishes PG9 and PG16 from monoclonal antibody 2909, and accounts for the neutralization breadth of PG9 and PG16, is their dependence on the asparagine-linked carbohydrate moiety that is present in most HIV-1 isolates at position 160 in V2 (Fig. 2b,c). The neutralization breadth of PG9 and PG16 indicates that the epitopes they target are conserved and not masked in most strains of HIV-1.
The immunological reactivity and neutralizing activity of the human QNE-specific monoclonal antibodies show that they can readily tolerate extensive amino acid changes in V3 (Refs 52, 53), and that broad or narrow neutralization activity depends on V2 variation. Furthermore, the broad neutralizing activity of PG9 and PG16 shows that the quaternary epitopes they recognize are composed of conserved elements in the V2 and V3 sequence-variable regions and are some of the most accessible of all neutralizing epitopes present on the CD4-unliganded form of the trimeric Env spike. The modelled Env trimers63,64,65 provide clues for visualizing the 3D structures of V2 and V3 contacts, which probably occur within the many possible unbound forms of the Env trimer. The resolved 3D structure (or structures) of Env trimers in QNE-bound conformations ultimately should be highly informative for vaccine design.
In summary, recent data reveal conserved targets for broadly reactive antibodies in the sequence-variable V2 and V3 loops of gp120 and on quaternary structures formed by these loops in the trimeric Env. These data explain the broad immunological and functional cross-reactivity of some of the monoclonal antibodies specific for these regions and provide a framework for conceptualizing the value of such regions in vaccine design.
Structural view of epitope mapping
Although masking is often described in terms of the ability of glycans and other structural features to prevent or decrease the neutralizing activity of V3-specific antibodies19,34,38,47,92, epitope masking is a phenomenon that is common to all HIV-1 neutralizing epitopes and is the result of many factors67. For example, the 3D structure of a particular epitope can assemble by adopting the 'correct' conformation that clusters the key antibody-binding amino acid side-chains together into a single structure, but the protein may be designed to easily 'flicker' out of that conformation, which would be observed as masking of the epitope from its cognate antibody. Similarly, other protein loops or glycans might 'flicker' in to cover an epitope, which would also be observed immunologically as partial or complete masking. The result is that, although antibodies might be elicited by immunization strategies targeting epitopes that are broadly present in circulating viruses, the effective neutralization mediated by these antibodies in vivo is decreased to an uncertain extent by different types of masking.
Masking can sometimes be overcome when particular epitope conformations are stabilized by various molecular ligands. For example, in some strains of HIV-1, V3 epitope accessibility on gp120 is markedly increased by binding to CD4 and certain gp120-specific monoclonal antibodies that induce and stabilize conformational changes in gp120 (Refs 39, 93, 94). These data suggest that exposure of 'masked' epitopes could be accomplished, and effective neutralizing activity thereby achieved, by inducing 'unmasking' antibodies together with antibodies specific for the 'masked' epitope (or epitopes). Another view of masking is illustrated by some monoclonal antibodies that have different neutralizing activities despite the fact that they target similar regions of Env (Fig. 2c). Binding of V2-, V3- and QNE-specific antibodies to overlapping regions of V2 and V3 has different effects that might depend on the molecular dynamics of V2 and V3 loops in different conformations of the Env trimer and/or on the kinetics of V2 and V3 movement as they flicker between low- and high-energy configurations95.
Masking might, therefore, be a result of the functional, as well as the physical, characteristics of an epitope in any of several molecular conformations. Hypothetically, residues in V2 and V3 could form the epitope (or epitopes) recognized by QNE-specific antibodies only when V2 and V3 interact in a configuration that leads to virus infectivity. By contrast, some individual V2- and V3-specific antibodies might only recognize the epitopes on their respective loops when these loops occupy different space — that is, are not in contact with one another — as in trimeric Env spikes that are not in an infectious conformation. The epitopes that are not in contact with one another would seem to be 'masked', as these V2- and V3-specific antibodies cannot neutralize in this case.
Attempts to understand masking fully probably require interdisciplinary studies that include structural, as well as immunological and virological, elements.
Conclusions and the way forward
Although the HIV-1 Env glycoproteins use variation — in terms of both sequence and conformation — to evade neutralization, there are nevertheless regions of structural conservation that can be targeted for vaccine design. This view is supported by data showing that sequence-variable loops of HIV-1 gp120 can elicit a range of antibodies, some with narrow specificities for only one or a few strains of HIV-1, and others with broad immunological and neutralizing activities against diverse HIV-1 strains. This is particularly true of antibodies specific for V2, V3 and the QNEs formed by these two regions of gp120. The immunological cross-reactivity of many of the monoclonal antibodies specific for these epitopes can now be explained in terms of the structural conservation of 3D shapes within these sequence-variable loops and in the context of the functional requirements that limit the quality and quantity of sequence variability. This synthesis of the structural, functional and antigenic conservation of sequence-variable regions of gp120 provides a paradigm for the rational design of recombinant immunogens: it shows how form must follow function, it recalls how form relates to antigenicity and it suggests how immunogens can be rationally designed to focus the immune response towards the portions of structures that elicit broadly reactive antibodies and away from those regions that elicit antibodies with narrow reactivity. Such rationally designed immunogens will be modelled on the detailed 3D structures of epitopes that are recognized by families of cross-clade and broadly neutralizing antibodies, targeting conserved structural features and eliminating or replacing those elements that narrow the antibody specificity.
New techniques have led to a large increase in the number and specificities of monoclonal antibodies that can be derived from patient specimens19,20,51,53,73. Other newly discovered epitopes and as yet unidentified epitopes recognized by serum antibodies with HIV-1 neutralizing activity10,73,74,96,97,98 will provide additional vaccine targets beyond those that have been described here. Using structural and computational biology to rationally design recombinant epitope–scaffold immunogens based on neutralizing epitopes in the variable regions of Env has the potential of developing immunogens that elicit antibodies with greater specificity and potency than are currently achieved when various forms of gp120 and/or gp41 are used. Indeed, until recently, there has been limited success in inducing broadly reactive polyclonal neutralizing antibody responses that target selected epitopes in gp120 and gp41 (Refs 6, 7, 8, 9 and summarized in Ref. 99). However, focusing the immune response on neutralizing epitopes through the use of epitope–scaffold vaccines designed to present conserved structural elements to the immune system and to eliminate those epitopes that narrow antibody specificity is beginning to yield modest successes44. These techniques hold the promise of constructing immunogens that elicit polyclonal antibody responses specific for particular chosen epitopes of HIV-1 with finer specificity and greater potency than is currently achieved.
References
Gilbert, P. B. et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191, 666–677 (2005).
Buchbinder, S. P. et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372, 1881–1893 (2008).
Rerks-Ngarm, S. et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361, 2209–2220 (2009).
[No authors listed.] Understanding antibody functions: beyond neutralization. IAVI Report (ed. Primer, V.) (2010).
Starcich, B. R. et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648 (1986).
Selvarajah, S. et al. Comparing antigenicity and immunogenicity of engineered gp120. J. Virol. 79, 12148–12163 (2005).
Selvarajah, S. et al. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res. Hum. Retroviruses 24, 301–314 (2008).
Law, M., Cardoso, R. M., Wilson, I. A. & Burton, D. R. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime–protein boost immunization strategy. J. Virol. 81, 4272–4285 (2007).
Saphire, E. O. et al. Structure of a high-affinity “mimotope” peptide bound to HIV-1-neutralizing antibody b12 explains its inability to elicit gp120 cross-reactive antibodies. J. Mol. Biol. 369, 696–709 (2007).
Li, Y. et al. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83, 1045–1059 (2009).
Simek, M. D. et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83, 7337–7348 (2009).
Kabat, E. A. & Mayer, M. M. Experimental Immunochemistry (ed. Charles, C.) (Thomas, Springfield, USA, 1961).
Israel, Z. R., Gorny, M. K., Palmer, C., McKeating, J. A. & Zolla-Pazner, S. Prevalence of a V2 epitope in clade B primary isolates and its recognition by sera from HIV-1 infected individuals. AIDS 11, 128–130 (1997).
Dalgleish, A. G. et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767 (1984).
Feng, Y., Broder, C. C., Kennedy, P. E. & Berger, E. A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science 272, 872–877 (1996).
Rizzuto, C. D. et al. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280, 1949–1953 (1998).
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Roben, P. et al. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68, 4821–4828 (1994).
Corti, D. et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE 5, e8805 (2010).
Wu, X. et al. Rational design of envelope surface identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science (in the press).
Thali, M. et al. Characterization of conserved HIV-type 1 gp120 neutralization epitopes exposed upon gp120–CD4 binding. J. Virol. 67, 3978–3988 (1993).
Zhang, M. Y., Shu, Y., Sidorov, I. & Dimitrov, D. S. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res. 61, 161–164 (2004).
Moulard, M. et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120–CD4–CCR5 complexes. Proc. Natl Acad. Sci. USA 99, 6913–6918 (2002).
Zhang, M. Y. & Dimitrov, D. S. Novel approaches for identification of broadly cross-reactive HIV-1 neutralizing human monoclonal antibodies and improvement of their potency. Curr. Pharm. Des. 13, 203–212 (2007).
Labrijn, A. F. et al. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77, 10557–10565 (2003).
Shioda, T., Levy, J. A. & Cheng-Mayer, C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA 89, 9434–9438 (1992).
Labrosse, B., Treboute, C., Brelot, A. & Alizon, M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 75, 5457–5464 (2001).
Trkola, A. et al. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384, 184–187 (1996).
Hill, C. M. et al. Envelope glycoproteins from HIV-1, HIV-2 and SIV can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71, 6296–6304 (1997).
Sharon, M. et al. Alternative conformations of HIV-1 V3 loops mimic β hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure (Camb.) 11, 225–236 (2003).
Cardozo, T. et al. Structural basis of co-receptor selectivity by the HIV-1 V3 loop. AIDS Res. Hum. Retroviruses 23, 415–426 (2007).
Gaschen, B. et al. Diversity considerations in HIV-1 vaccine selection. Science 296, 2354–2360 (2002).
Cao, J. et al. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71, 9808–9812 (1997).
Davis, K. L. et al. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387, 414–426 (2009).
Li, M. et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79, 10108–10125 (2005).
Pantophlet, R., Aguilar-Sino, R. O., Wrin, T., Cavacini, L. A. & Burton, D. R. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology 364, 441–453 (2007).
Hioe, C. E. et al. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE 5, e10254 (2010).
Bou-Habib, D. C. et al. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68, 6006–6013 (1994).
Wu, X. et al. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology 380, 285–295 (2008).
Almond, D. et al. Structural conservation predominates over sequence variability in the crown of HIV-1's V3 loop. AIDS Res. Hum. Retroviruses (in the press).
Jiang, X. et al. Conserved structural elements in the V3 crown of HIV-1 gp120. Nature Struct. Mol. Biol. (in the press).
Zolla-Pazner, S. Improving on nature: focusing the immune response on the V3 Loop. Hum. Antibodies 14, 69–72 (2005).
Gorny, M. K. et al. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of HIV-1. J. Virol. 80, 6865–6872 (2006).
Zolla-Pazner, S. et al. Cross-clade neutralizing antibodies against HIV-1 induced in rabbits by focusing the immune response on a neutralizing epitope. Virology 392, 82–93 (2009).
Arthos, J. et al. HIV-1 envelope protein binds to and signals through integrin α4β7, the gut mucosal homing receptor for peripheral T cells. Nature Immunol. 9, 301–309 (2008).
Chen, B. et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841 (2005).
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003).
Sagar, M., Wu, X., Lee, S. & Overbaugh, J. Human immunodeficiency virus type 1 V1–V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80, 9586–9598 (2006).
Derdeyn, C. A. et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303, 2019–2022 (2004).
Ly, A. & Stamatatos, L. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74, 6769–6776 (2000).
Gorny, M. K. et al. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79, 5232–5237 (2005).
Honnen, W. J. et al. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81, 1424–1432 (2007).
Walker, L. M. et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 (2009).
Robinson, J. E. et al. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4 . J. Virol. 84, 3443–3453 (2010).
He, Y. et al. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J. Immunol. 169, 595–605 (2002).
Huang, Z. et al. Levels of N-linked glycosylation on the V1 loop of HIV-1 Env proteins and their relationship to the antigenicity of Env from primary viral isolates. Curr. HIV Res. 6, 296–305 (2008).
Sullivan, N., Thali, M., Furman, C., Ho, D. D. & Sodroski, J. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67, 3674–3679 (1993).
Moore, P. L. et al. The C3–V4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 82, 1860–1869 (2008).
Burke, B. et al. Neutralizing antibody responses to subtype B and C adjuvanted HIV envelope protein vaccination in rabbits. Virology 387, 147–156 (2009).
Dieltjens, T. et al. HIV type 1 subtype A envelope genetic evolution in a slow progressing individual with consistent broadly neutralizing antibodies. AIDS Res. Hum. Retroviruses 25, 1165–1169 (2009).
Castro, E. et al. Independent evolution of hypervariable regions of HIV-1 gp120: V4 as a swarm of N-linked glycosylation variants. AIDS Res. Hum. Retroviruses 24, 106–113 (2008).
Rong, R. et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5, e1000594 (2009).
Zanetti, G., Briggs, J. A., Grunewald, K., Sattentau, Q. J. & Fuller, S. D. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2, 790–797 (2006).
Zhu, P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 (2006).
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Wyatt, R. et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393, 705–711 (1998).
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002).
Kimura, T., Wang, X. H., Williams, C., Zolla-Pazner, S. & Gorny, M. K. Human monoclonal antibody 2909 binds to pseudovirions expressing trimers but not monomeric HIV-1 envelope proteins. Hum. Antibodies 18, 35–40 (2009).
Gorny, M. K., Xu, J.-Y., Karwowska, S., Buchbinder, A. & Zolla-Pazner, S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150, 635–643 (1993).
Burke, V. et al. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 monoclonal antibodies: implications for design of V3-based immunogens. Structure 17, 1538–1546 (2009).
Conley, A. J. et al. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68, 6994–7000 (1994).
Binley, J. M. et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78, 13232–13252 (2004).
Scheid, J. F. et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640 (2009).
Nandi, A. et al. Epitopes for broad and potent neutralizing antibody responses during chronic infection with human immunodeficiency virus type 1. Virology 396, 339–348 (2010).
Pantophlet, R., Wrin, T., Cavacini, L. A., Robinson, J. E. & Burton, D. R. Neutralizing activity of antibodies to the V3 loop region of HIV-1 gp120 relative to their epitope fine specificity. Virology 381, 251–260 (2008).
Stanfield, R. L., Gorny, M. K., Williams, C., Zolla-Pazner, S. & Wilson, I. A. Structural rationale for the broad neutralization of HIV-1 by human antibody 447-52D. Structure 12, 193–204 (2004).
Stanfield, R. L., Gorny, M. K., Zolla-Pazner, S. & Wilson, I. A. Crystal structures of HIV-1 neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J. Virol. 80, 6093–6105 (2006).
Dhillon, A. K. et al. Structure determination of an anti-HIV-1 Fab 447-52D–peptide complex from an epitaxially twinned data set. Acta Crystallogr. D Biol. Crystallogr. 64, 792–802 (2008).
Bell, C. H. et al. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J. Mol. Biol. 375, 969–978 (2008).
Huang, C. C. et al. Structure of a V3-containing HIV-1 gp120 core. Science 310, 1025–1028 (2005).
Wu, Z. et al. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J. Virol. 69, 2271–2278 (1995).
McKeating, J. A. et al. Immunogenicity of full length and truncated forms of the human immunodeficiency virus type I envelope glycoprotein. Immunol. Lett. 51, 101–105 (1996).
Gorny, M. K. et al. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of HIV-1. J. Virol. 68, 8312–8320 (1994).
Moore, J. P. et al. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J. Virol. 67, 6136–6151 (1993).
Warrier, S. V. et al. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68, 4636–4642 (1994).
Pinter, A. et al. The V1/V2 domain of gp120 is a global regulator of sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78, 5205–5215 (2004).
Atkinson, H. J. & Babbitt, P. C. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput. Biol. 5, e1000541 (2009).
Stamatatos, L. & Cheng-Mayer, C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72, 7840–7845 (1998).
Etemad-Moghadam, B. et al. Characterization of simian–human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected monkeys. J. Virol. 72, 8437–8445 (1998).
Chen, C. H., Jin, L., Zhu, C., Holz-Smith, S. & Matthews, T. J. Induction and characterization of neutralizing antibodies against a human immunodeficiency virus type 1 primary isolate. J. Virol. 75, 6700–6704 (2001).
Cho, M. W., Lee, M. K., Chen, C. H., Matthews, T. & Martin, M. A. Identification of gp120 regions targeted by a highly potent neutralizing antiserum elicited in a chimpanzee inoculated with a primary human immunodeficiency virus type 1 isolate. J. Virol. 74, 9749–9754 (2000).
Krachmarov, C. et al. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80, 7127–7135 (2006).
Mbah, H. A. et al. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J. Virol. 75, 7785–7788 (2001).
Hioe, C. E. et al. The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120. Vaccine 28, 352–360 (2009).
Hsu, S. T. & Bonvin, A. M. Atomic insight into the CD4 binding-induced conformational changes in HIV-1 gp120. Proteins 55, 582–593 (2004).
Guan, Y. et al. Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc. Natl Acad. Sci. USA 106, 3952–3957 (2009).
Binley, J. M. et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82, 11651–11668 (2008).
Sather, D. N. et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83, 757–769 (2009).
Arnold, G. F. et al. Broad neutralization of human immunodeficiency virus type 1 (HIV-1) elicited from human rhinoviruses that display the HIV-1 gp41 ELDKWA epitope. J. Virol. 83, 5087–5100 (2009).
Gorny, M. K. et al. Neutralization of diverse HIV-1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66, 7538–7542 (1992).
Hochleitner, E. O., Gorny, M. K., Zolla-Pazner, S. & Tomer, K. B. Mass spectrometric characterization of a discontinuous epitope of the HIV envelope protein HIV-gp120 recognized by the human monoclonal antibody 1331A. J. Immunol. 164, 4156–4161 (2000).
Robinson, W. E. Jr, Gorny, M. K., Xu, J. Y., Mitchell, W. M. & Zolla-Pazner, S. Two immunodominant domains of gp41 bind antibodies which enhance HIV-1 infection in vitro. J. Virol. 65, 4169–4176 (1991).
Gorny, M. K. & Zolla-Pazner, S. Recognition of free and complexed peptides representing the prefusogenic and fusogenic forms of HIV-1 gp41 by human monoclonal antibodies. J. Virol. 74, 6186–6192 (2000).
Zhou, T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007).
Moore, J. P. & Sodroski, J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70, 1863–1872 (1996).
Acknowledgements
We are grateful to D. Almond and J. Swetnam for assistance in preparing the figures and to C. Hioe, M. Totrov and X.-P. Kong for contributing ideas to and helping to shape this article, which includes studies supported by grants from the Bill and Melinda Gates Foundation, the US National Institutes of Health (grants HL59725, AI38065, DP2 OD004631 and AI084119) and the US Department of Veterans Affairs.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Zolla-Pazner, S., Cardozo, T. Structure–function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol 10, 527–535 (2010). https://doi.org/10.1038/nri2801
Issue Date:
DOI: https://doi.org/10.1038/nri2801
This article is cited by
-
A voting mechanism-based linear epitope prediction system for the host-specific Iridoviridae family
BMC Bioinformatics (2019)
-
Analysis of HIV-1 envelope evolution suggests antibody-mediated selection of common epitopes among Chinese former plasma donors from a narrow-source outbreak
Scientific Reports (2018)
-
Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer
Nature Communications (2018)
-
The CpG dinucleotide content of the HIV-1 envelope gene may predict disease progression
Scientific Reports (2017)
-
Conformational Heterogeneity of the HIV Envelope Glycan Shield
Scientific Reports (2017)