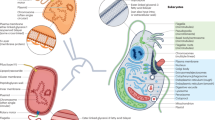
Key Points
-
Horizontal gene transfer (HGT; also known as lateral gene transfer) is the non-sexual movement of genetic information between genomes. Incoming DNA or RNA can replace existing genes, or can introduce new genes into a genome.
-
HGT in eukaryotes has been overshadowed by its prevalence in bacterial genomes, but a large number of cases involving eukaryotes have nevertheless been described. Its impact is variable from lineage to lineage, and many of the lineages that are most affected by HGT are the least studied at the genomic level (for example, protists), so the importance of HGT will probably increase as databases expand.
-
There is an extensive history of endosymbiosis in eukaryotic evolution. Endosymbiosis-derived organelles like mitochondria and plastids contributed many genes to the nucleus, and subsequent endosymbioses in plastid evolution has also allowed genes to move between eukaryotes. Other persistent endosymbionts (for example, Wolbachia) have now been recognized as a major source of DNA in the nuclei of their hosts.
-
HGT without symbiosis is also common, most documented cases involve genes from bacteria being present in the nucleus. Their prevalence might be because they are more obvious than genes from another eukaryote, but bacteria might be the major source of new genes owing to their abundance.
-
The complex distribution of some previously 'simple' bacteria–eukaryote transfers suggests eukaryote–eukaryote transfers might be more common than is appreciated. Extensive exchange of genetic information between mitochondrial genomes of plants is now well documented, providing the highest known levels of eukaryote–eukaryote transfer.
-
In many instances, genes acquired by HGT have clear functional or ecological implications for their new host. For example, several highly derived anaerobes have acquired many metabolic genes from bacteria that probably contributed to their adaptation to anaerobic environments.
-
Although the confounding effect of HGT on phylogenetic reconstruction is well known, ancient HGT events have been recognized as useful clues to organismal relationships. Several ancient HGT events are shared by two or more major eukaryotic lineages, such as animals and fungi, or chromalveolates, and such shared characters (interpreted with caution) help understand the tree of eukaryotes.
-
Recent transfers are observed more often than ancient ones. This might be a sampling bias, but could indicate that transferred genes provide a transient advantage but are frequently not retained over prolonged periods of time. If so, the impact of HGT in the long term might not be so profound.
-
Even with the growth of genomic data, several questions remain challenging: the fate of transferred genes over time, the mechanisms of HGT, and what dictates the impact of HGT on different lineages are all major outstanding questions. On the basis of the history of HGT research in bacteria, it seems likely that many questions that seem simple now will become more complex as data accumulates.
Abstract
Horizontal gene transfer (HGT; also known as lateral gene transfer) has had an important role in eukaryotic genome evolution, but its importance is often overshadowed by the greater prevalence and our more advanced understanding of gene transfer in prokaryotes. Recurrent endosymbioses and the generally poor sampling of most nuclear genes from diverse lineages have also complicated the search for transferred genes. Nevertheless, the number of well-supported cases of transfer from both prokaryotes and eukaryotes, many with significant functional implications, is now expanding rapidly. Major recent trends include the important role of HGT in adaptation to certain specialized niches and the highly variable impact of HGT in different lineages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Doolittle, W. F. Phylogenetic classification and the universal tree. Science 284, 2124–2129 (1999).
Eisen, J. A. Horizontal gene transfer among microbial genomes: new insights from complete genome analysis. Curr. Opin. Genet. Dev. 10, 606–611 (2000).
Koonin, E. V., Makarova, K. S. & Aravind, L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55, 709–742 (2001).
Kurland, C. G. What tangled web: barriers to rampant horizontal gene transfer. Bioessays 27, 741–747 (2005).
Ochman, H., Lawrence, J. G. & Groisman, E. A. Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (2000).
Lawrence, J. G. Gene transfer in bacteria: speciation without species? Theor. Popul. Biol. 61, 449–460 (2002).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Salzberg, S. L., White, O., Peterson, J. & Eisen, J. A. Microbial genes in the human genome: lateral transfer or gene loss? Science 292, 1903–1906 (2001).
Stanhope, M. J. et al. Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature 411, 940–944 (2001).
Andersson, J. O. in Genomics and Evolution of Microbial Eukaryotes (eds Katz, L. A. & Bhattacharya, D.) 109–122 (Oxford Univ. Press, Oxford, 2006).
Andersson, J. O. Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197 (2005).
Richards, T. A., Hirt, R. P., Williams, B. A. & Embley, T. M. Horizontal gene transfer and the evolution of parasitic protozoa. Protist 154, 17–32 (2003).
Ragan, M. A., Harlow, T. J. & Beiko, R. G. Do different surrogate methods detect lateral genetic transfer events of different relative ages? Trends Microbiol. 14, 4–8 (2006).
Roelofs, J. & Van Haastert, P. J. Genes lost during evolution. Nature 411, 1013–1014 (2001).
Roger, A. J. Reconstructing early events in eukaryotic evolution. Am. Nat. 154, S146–S163 (1999).
Andersson, J. O., Hirt, R. P., Foster, P. G. & Roger, A. J. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol. Biol. 6, 27 (2006). Even among just four gene families, a large number of both bacteria–eukaryote and eukaryote–eukaryote transfers are shown to have accumulated sequentially in the genomes of several diverse lineages of protists.
Noble, G. P., Rogers, M. B. & Keeling, P. J. Complex distribution of EFL and EF-1alpha proteins in the green algal lineage. BMC Evol. Biol. 7, 82 (2007). The overall distribution of the EFL protein suggests several eukaryote–eukaryote HGT events (see reference 87), but the distribution of EFL within green algae is so complex at the fine scale that both transfer and lineage sorting probably contributed to its evolution.
Stanier, R. Y. Some aspects of the biology of cells and their possible evolutionary significance. Symp. Soc. Gen. Microbiol. 20, 1–38 (1970).
Hoffmeister, M. & Martin, W. Interspecific evolution: microbial symbiosis, endosymbiosis and gene transfer. Environ. Microbiol. 5, 641–649 (2003).
de Souza, W. & Motta, M. C. Endosymbiosis in protozoa of the Trypanosomatidae family. FEMS Microbiol. Lett. 173, 1–8 (1999).
Hackstein, J. H. & Vogels, G. D. Endosymbiotic interactions in anaerobic protozoa. Antonie Van Leeuwenhoek 71, 151–158 (1997).
Wernegreen, J. J. Endosymbiosis: lessons in conflict resolution. PLoS Biol. 2, e68 (2004).
Slamovits, C. H., Saldarriaga, J. F., Larocque, A. & Keeling, P. J. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 372, 356–368 (2007).
Martin, W. et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl Acad. Sci. USA 99, 12246–12251 (2002).
Kurland, C. G. & Andersson, S. G. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64, 786–820 (2000).
Obornik, M. & Green, B. R. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 22, 2343–2353 (2005).
Richards, T. A. et al. Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5, 1517–1531 (2006).
Patron, N. J. & Keeling, P. J. Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J. Phycol. 41, 1131–1141 (2005).
Esser, C. et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004).
Adams, K. L. & Palmer, J. D. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380–395 (2003).
Millen, R. S. et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658 (2001).
Adams, K. L., Qiu, Y. L., Stoutemyer, M. & Palmer, J. D. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl Acad. Sci. USA 99, 9905–9912 (2002). A large-scale study showing that rates of functional transfer of mitochondrial genes to the nucleus in angiosperm vary enormously, often exceeding rates of even synonymous nucleotide substitutions, and that genes for hydrophobic proteins are far more recalcitrant to functional transfer than are those encoding non-hydrophobic proteins.
Adams, K. L., Daley, D. O., Qiu, Y. L., Whelan, J. & Palmer, J. D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357 (2000).
Huang, C. Y., Ayliffe, M. A. & Timmis, J. N. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422, 72–76 (2003).
Thorsness, P. E. & Weber, E. R. Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int. Rev. Cytol. 165, 207–234 (1996).
Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nature Rev. Genet. 5, 123–135 (2004).
Leister, D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 21, 655–663 (2005).
Matsuo, M., Ito, Y., Yamauchi, R. & Obokata, J. The rice nuclear genome continuously integrates, shuffles, and eliminates the chloroplast genome to cause chloroplast–nuclear DNA flux. Plant Cell 17, 665–675 (2005). Elegant bioinformatic analyses reveal that, in rice at least, plastid DNA sequences are transferred to the nucleus at even higher rates than previously observed but are then lost and fragmented at surprisingly high rates too. Another unexpected finding is that much of this chloroplast–nuclear flux occurs in pericentromeric regions of chromosomes.
Henze, K. & Martin, W. How do mitochondrial genes get into the nucleus? Trends Genet. 17, 383–387 (2001).
Choi, C., Liu, Z. & Adams, K. L. Evolutionary transfers of mitochondrial genes to the nucleus in the Populus lineage and coexpression of nuclear and mitochondrial Sdh4 genes. New Phytol. 172, 429–439 (2006).
van Dooren, G. G., Su, V., D'Ombrain, M. C. & McFadden, G. I. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J. Biol. Chem. 277, 23612–23619 (2002).
Hotopp, J. C. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007). Many animals harbour bacterial endosymbionts, and this paper shows that many genes, or even whole copies of these endosymbiont genomes, have been transferred to the nuclear genome of the animal host. This paper is also a cautionary tale for the automated removal of 'contaminant' bacterial sequence from genome projects of eukaryotes that harbour bacterial symbionts.
Nikoh, N. et al. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 18, 272–280 (2008).
Nakabachi, A. et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314, 267 (2006).
Tamames, J. et al. The frontier between cell and organelle: genome analysis of Candidatus Carsonella ruddii. BMC Evol. Biol. 7, 181 (2007).
McCutcheon, J. P. & Moran, N. A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl Acad. Sci. USA 104, 19392–19397 (2007).
Dujon, B. et al. Genome evolution in yeasts. Nature 430, 35–44 (2004).
Hall, C., Brachat, S. & Dietrich, F. S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4, 1102–1115 (2005).
Ricard, G. et al. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7, 22 (2006). The rumen gut is an environment that is rich in bacteria–protist interactions, and this study shows that rumen ciliates are taking up bacterial genes at a high frequency.
Doolittle, W. F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311 (1998).
Archibald, J. M., Rogers, M. B., Toop, M., Ishida, K. & Keeling, P. J. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl Acad. Sci. USA 100, 7678–7683 (2003). One of the first studies to describe widespread HGT to a nuclear genome, in this case showing that plastid-targeted genes are derived from many different kinds of algae and bacteria.
Watkins, R. F. & Gray, M. W. The frequency of eubacterium-to-eukaryote lateral gene transfers shows significant cross-taxa variation within amoebozoa. J. Mol. Evol. 63, 801–814 (2006).
de Koning, A. P., Brinkman, F. S., Jones, S. J. & Keeling, P. J. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 17, 1769–1773 (2000).
Andersson, J. O., Doolittle, W. F. & Nesbo, C. L. Genomics. Are there bugs in our genome? Science 292, 1848–1850 (2001).
Kondrashov, F. A., Koonin, E. V., Morgunov, I. G., Finogenova, T. V. & Kondrashova, M. N. Evolution of glyoxylate cycle enzymes in metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 1, 31 (2006).
Nakashima, K., Yamada, L., Satou, Y., Azuma, J. & Satoh, N. The evolutionary origin of animal cellulose synthase. Dev. Genes Evol. 214, 81–88 (2004).
Berriman, M. et al. The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 (2005).
Douzery, E. J., Snell, E. A., Bapteste, E., Delsuc, F. & Philippe, H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl Acad. Sci. USA 101, 15386–15391 (2004).
Andersson, J. O., Sarchfield, S. W. & Roger, A. J. Gene transfers from nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol. Biol. Evol. 22, 85–90 (2005).
Andersson, J. O. et al. A genomic survey of the fish parasite Spironucleus salmonicida indicates genomic plasticity among diplomonads and significant lateral gene transfer in eukaryote genome evolution. BMC Genomics 8, 51 (2007). Dozens of putative HGTs were identified in the genome of one diplomonad, with most of these events pre-dating the divergence of the two major lineages of diplomonads and some apparently pre-dating the divergence of diplomonads and parabasalians. Most transferred genes encode metabolic proteins and were acquired from bacteria, but a significant minority seem to be eukaryote-to-eukaryote transfers.
Derelle, E. et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl Acad. Sci. USA 103, 11647–11652 (2006).
Andersson, J. O., Sjogren, A. M., Davis, L. A., Embley, T. M. & Roger, A. J. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 13, 94–104 (2003).
Slamovits, C. H. & Keeling, P. J. Pyruvate-phosphate dikinase of oxymonads and parabasalia and the evolution of pyrophosphate-dependent glycolysis in anaerobic eukaryotes. Eukaryot. Cell 5, 148–154 (2006).
Carlton, J. M. et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315, 207–212 (2007).
Morrison, H. G. et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317, 1921–1926 (2007).
Loftus, B. et al. The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868 (2005). Genome sequencing of anaerobic eukaryotic parasites (also see references 64 and 65) has confirmed the conclusion, which was based on earlier, single-gene studies, that HGT has had a major role in the ad hoc assembly of the unusual metabolic pathways of these organisms.
Eichinger, L. et al. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 (2005).
Waller, R. F., Slamovits, C. H. & Keeling, P. J. Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 23, 1437–1443 (2006).
Andersson, J. O. & Roger, A. J. Evolutionary analyses of the small subunit of glutamate synthase: gene order conservation, gene fusions, and prokaryote-to-eukaryote lateral gene transfers. Eukaryot. Cell 1, 304–310 (2002).
Rice, D. W. & Palmer, J. D. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4, 31 (2006). In contrast to plant mitochondria, this large-scale analysis of HGT in plastid genomes revealed very little transfer. One case that was identified, however, is an important, rare character that serves to phylogenetically unite two major groups of algae.
Khan, H. et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol. Biol. Evol. 24, 1832–1842 (2007).
Bevan, R. B. & Lang, B. F. in Topics in Current Genetics Vol. 8. (eds Kohler, C. & Bauer, M. F.) 1–35 (Springer, Berlin, 2004).
Bachvaroff, T. R., Concepcion, G. T., Rogers, C. R., Herman, E. M. & Delwiche, C. F. Dinoflagellate expressed sequence tags data indicate massive trasfer of chloroplast genes to the nuclear genome. Protist 155, 65–78 (2004).
Keeling, P. J. & Palmer, J. D. Lateral transfer at the gene and subgenic levels in the evolution of eukaryotic enolase. Proc. Natl Acad. Sci. USA 98, 10745–10750 (2001).
Patron, N. J., Waller, R. F. & Keeling, P. J. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 357, 1373–1382 (2006). Some dinoflagellates have a complex history of replacing their plastids with new ones from other lineages. Here it is shown that genes derived from both plastids can be retained in the nuclear genome and their products also targeted to the new plastid, making its proteome chimeric.
Rogers, M. B. & Keeling, P. J. Lateral gene transfer and re-compartmentalisation of Calvin cycle enzymes in plants and algae. J. Mol. Evol. 58, 367–375 (2003).
Hackett, J. D. et al. Migration of the plastid genome to the nucleus in a peridinin dinoflagellate. Curr. Biol. 14, 213–218 (2004).
Waller, R. F., Patron, N. J. & Keeling, P. J. Phylogenetic history of plastid-targeted proteins in the peridinin-containing dinoflagellate Heterocapsa triquetra. Int. J. Syst. Evol. Microbiol. 56, 1439–1347 (2006).
Richards, T. A., Dacks, J. B., Jenkinson, J. M., Thornton, C. R. & Talbot, N. J. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16, 1857–1864 (2006). Fungal and oomycete plant pathogens are distantly related but have many mechanistic similarities, and here it is shown that several convergent characteristics of their pathogenicity and feeding might be due to transfer of genes between them.
Friesen, T. L. et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nature Genet. 38, 953–956 (2006). Virulence-related factors have been observed to transfer in several systems, including fungi, but in this case a virulence factor has moved from one fungus to another so recently that it can be dated to around 1941, when it led to the emergence of a new disease.
Inderbitzin, P., Harkness, J., Turgeon, B. G. & Berbee, M. L. Lateral transfer of mating system in Stemphylium. Proc. Natl Acad. Sci. USA 102, 11390–11395 (2005).
Slot, J. C. & Hibbett, D. S. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS ONE 2, e1097 (2007).
Simpson, A. G., Perley, T. A. & Lara, E. Lateral transfer of the gene for a widely used marker, alpha-tubulin, indicated by a milti-protein study of the phylogenetic position of Andalucia (Excavata). Mol. Phylogenet. Evol. 47, 366–377 (2008).
Paoletti, M., Buck, K. W. & Brasier, C. M. Selective acquisition of novel mating type and vegetative incompatibility genes via interspecies gene transfer in the globally invading eukaryote Ophiostoma novo-ulmi. Mol. Ecol. 15, 249–262 (2006).
Hall, C. & Dietrich, F. S. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177, 2293–2307 (2007).
Kavanaugh, L. A., Fraser, J. A. & Dietrich, F. S. Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol. Biol. Evol. 23, 1879–1890 (2006).
Keeling, P. J. & Inagaki, Y. A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1alpha. Proc. Natl Acad. Sci. USA 101, 15380–15385 (2004).
Rogers, M. B. et al. A complex and punctate distribution of three eukaryotic genes derived by lateral gene transfer. BMC Evol. Biol. 7, 89 (2007). There are many examples of relatively simple bacteria–eukaryote HGT events, but in this paper broader sampling of three cases shows that complex, multiple transfer events might often seem to be simpler than they really are, and that transfer between eukaryotes might be more common than we think.
Nosenko, T. & Bhattacharya, D. Horizontal gene transfer in chromalveolates. BMC Evol. Biol. 7, 173 (2007).
Alvarez, N. et al. Phylogeographic support for horizontal gene transfer involving sympatric bruchid species. Biol. Direct 1, 21 (2006).
Jenkins, C. et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl Acad. Sci. USA 99, 17049–17054 (2002). Tubulins are hallmark proteins of the eukaryotic cytoskeleton, so it was surprising to find that this bacterial genome had acquired both alpha- and beta-tubulin genes by HGT, and that the proteins seem to retain some structural function.
Schlieper, D., Oliva, M. A., Andreu, J. M. & Lowe, J. Structure of bacterial tubulin BtubA/B: evidence for horizontal gene transfer. Proc. Natl Acad. Sci. USA 102, 9170–9175 (2005).
Sontag, C. A., Staley, J. T. & Erickson, H. P. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J. Cell Biol. 169, 233–238 (2005).
Guljamow, A. et al. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr. Biol. 17, R757–R759 (2007). Similar to reference 91, the cytoskeletal protein actin was thought to be unique to eukaryotes, but this cyanobacterium was found to have taken up genes for actin and actin accessory proteins, which are now used in a structural capacity.
Rogers, M. B., Patron, N. J. & Keeling, P. J. Horizontal transfer of a eukaryotic plastid-targeted protein gene to cyanobacteria. BMC Biol. 5, 26 (2007). Many eukaryotic genes are derived from the plastid endosymbiont, but here a eukaryotic gene that took over plastid function has also been transferred back to cyanobacteria, where it integrated into the genome adjacent to its bacterial analogue.
Gamieldien, J., Ptitsyn, A. & Hide, W. Eukaryotic genes in Mycobacterium tuberculosis could have a role in pathogenesis and immunomodulation. Trends Genet. 18, 5–8 (2002).
Kinsella, R. J. & McInerney, J. O. Eukaryotic genes in Mycobacterium tuberculosis? Possible alternative explanations. Trends Genet. 19, 687–689 (2003).
Huang, J. & Gogarten, J. P. Ancient horizontal gene transfer can benefit phylogenetic reconstruction. Trends Genet. 22, 361–366 (2006).
Stechmann, A., Baumgartner, M., Silberman, J. D. & Roger, A. J. The glycolytic pathway of Trimastix pyriformis is an evolutionary mosaic. BMC Evol. Biol. 6, 101 (2006).
Henze, K. et al. Unique phylogenetic relationships of glucokinase and glucosephosphate isomerase of the amitochondriate eukaryotes Giardia intestinalis, Spironucleus barkhanus and Trichomonas vaginalis. Gene 281, 123–131 (2001).
Ledent, V. & Vervoort, M. Comparative genomics of the class 4 histone deacetylase family indicates a complex evolutionary history. BMC Biol. 4, 24 (2006).
Patron, N. J., Inagaki, Y. & Keeling, P. J. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol. 17, 887–891 (2007).
Hackett, J. D. et al. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 24, 1702–1713 (2007).
Huang, J., Xu, Y. & Gogarten, J. P. The presence of a haloarchaeal type tyrosyl-tRNA synthetase marks the opisthokonts as monophyletic. Mol. Biol. Evol. 22, 2142–2146 (2005).
Patron, N. J., Rogers, M. B. & Keeling, P. J. Gene replacement of fructose-1,6-bisphosphate aldolase (FBA) supports a single photosynthetic ancestor of chromalveolates. Eukaryot. Cell 3, 1169–1175 (2004).
Berney, C. & Pawlowski, J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc. Biol. Sci. 273, 1867–1872 (2006).
Hao, W. & Golding, G. B. Patterns of bacterial gene movement. Mol. Biol. Evol. 21, 1294–1307 (2004).
Hao, W. & Golding, G. B. The fate of laterally transferred genes: life in the fast lane to adaptation or death. Genome Res. 16, 636–643 (2006).
Jain, R., Rivera, M. C. & Lake, J. A. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801–3806 (1999).
Keeling, P. J. The diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 91, 1481–1493 (2004).
Reyes-Prieto, A., Weber, A. P. & Bhattacharya, D. The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 (2007).
Huang, J. & Gogarten, J. P. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8, R99 (2007).
Stiller, J. W. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 12, 391–396 (2007).
Archibald, J. M. Jumping genes and shrinking genomes — probing the evolution of eukaryotic photosynthesis with genomics. IUBMB Life 57, 539–447 (2005).
Ishida, K. & Green, B. R. Second- and third-hand chloroplasts in dinoflagellates: phylogeny of oxygen-evolving enhancer 1 (PsbO) protein reveals replacement of a nuclear-encoded plastid gene by that of a haptophyte tertiary endosymbiont. Proc. Natl Acad. Sci. USA 99, 9294–9299 (2002).
Cavalier-Smith, T. in Organelles, Genomes and Eukaryotic Evolution (ed. Horner, D.) 71–103 (Taylor and Francis, London, 2004).
Fast, N. M., Kissinger, J. C., Roos, D. S. & Keeling, P. J. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18, 418–426 (2001).
Abrahamsen, M. S. et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445 (2004).
Eisen, J. A. et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4, e286 (2006).
Tyler, B. M. et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 (2006).
Aury, J. M. et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444, 171–178 (2006).
Huang, J. et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 5, R88 (2004).
Moore, R. B. et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 451, 959–963 (2008).
Hannaert, V. et al. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl Acad. Sci. USA 100, 1067–1071 (2003).
El-Sayed, N. M. et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415 (2005).
Pfanner, N. & Geissler, A. Versatility of the mitochondrial protein import machinery. Nature Rev. Mol. Cell Biol. 2, 339–349 (2001).
Bhattacharya, D., Archibald, J. M., Weber, A. P. & Reyes-Prieto, A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29, 1239–1246 (2007).
Theissen, U. & Martin, W. The difference between organelles and endosymbionts. Curr. Biol. 16, R1016–1017; author reply R1017–R1018 (2006).
McFadden, G. I. Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 37, 951–959 (2001).
de Grey, A. D. Forces maintaining organellar genomes: is any as strong as genetic code disparity or hydrophobicity? Bioessays 27, 436–446 (2005).
Daley, D. O. & Whelan, J. Why genes persist in organelle genomes. Genome Biol. 6, 110 (2005). This is a balanced and insightful review of the many hypotheses to account for the persistence of certain genes in organellar genomes, with the importability hypothesis being the best-supported hypothesis.
Daley, D. O., Clifton, R. & Whelan, J. Intracellular gene transfer: reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc. Natl Acad. Sci. USA 99, 10510–10515 (2002).
Allen, J. F., Puthiyaveetil, S., Strom, J. & Allen, C. A. Energy transduction anchors genes in organelles. Bioessays 27, 426–435 (2005).
Burger, G., Gray, M. W. & Lang, B. F. Mitochondrial genomes: anything goes. Trends Genet. 19, 709–716 (2003).
Green, B. R. The chloroplast genome of dinoflagellates — a reduced instruction set? Protist 155, 23–31 (2004).
Funes, S. et al. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J. Biol. Chem. 277, 6051–6058 (2002).
Perez-Martinez, X. et al. Subunit II of cytochrome c oxidase in Chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J. Biol. Chem. 276, 11302–11309 (2001).
Richardson, A. O. & Palmer, J. D. Horizontal gene transfer in plants. J. Exp. Bot. 58, 1–9 (2007).
Koulintchenko, M., Konstantinov, Y. & Dietrich, A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 22, 1245–1254 (2003).
Arimura, S., Yamamoto, J., Aida, G. P., Nakazono, M. & Tsutsumi, N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl Acad. Sci. USA 101, 7805–7808 (2004).
Sheahan, M. B., McCurdy, D. W. & Rose, R. J. Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44, 744–755 (2005).
Mower, J. P., Stefanovic, S., Young, G. J. & Palmer, J. D. Plant genetics: gene transfer from parasitic to host plants. Nature 432, 165–166 (2004).
Won, H. & Renner, S. S. Horizontal gene transfer from flowering plants to Gnetum. Proc. Natl Acad. Sci. USA 100, 10824–10829 (2003).
Davis, C. C., Anderson, W. R. & Wurdack, K. J. Gene transfer from a parasitic flowering plant to a fern. Proc. Biol. Sci. 272, 2237–2242 (2005).
Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (2003).
Barkman, T. J. et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol. Biol. 7, 248 (2007).
Bergthorsson, U., Richardson, A. O., Young, G. J., Goertzen, L. R. & Palmer, J. D. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc. Natl Acad. Sci. USA 101, 17747–17752 (2004). A remarkable case in which a flowering plant mitochondrial genome possesses as many foreign genes, acquired from a diverse range of land plant mitochondrial donors, as it does native genes.
Nickrent, D. L., Blarer, A., Qiu, Y. L., Vidal-Russell, R. & Anderson, F. E. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol. Biol. 4, 40 (2004).
Davis, C. C. & Wurdack, K. J. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 305, 676–678 (2004).
Sanchez-Gracia, A., Maside, X. & Charlesworth, B. High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 21, 200–203 (2005).
Haugen, P., Simon, D. M. & Bhattacharya, D. The natural history of group I introns. Trends Genet. 21, 111–119 (2005).
Lambowitz, A. M. & Zimmerly, S. Mobile group II introns. Annu. Rev. Genet. 38, 1–35 (2004).
Cho, Y., Qiu, Y. L., Kuhlman, P. & Palmer, J. D. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl Acad. Sci. USA 95, 14244–14249 (1998).
Wei, W. et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl Acad. Sci. USA 104, 12825–12830 (2007).
Woolfit, M., Rozpedowska, E., Piskur, J. & Wolfe, K. H. Genome survey sequencing of the wine spoilage yeast Dekkera (Brettanomyces) bruxellensis. Eukaryot. Cell 6, 721–733 (2007).
Fukami, H., Chen, C. A., Chiou, C. Y. & Knowlton, N. Novel group I introns encoding a putative homing endonuclease in the mitochondrial cox1 gene of scleractinian corals. J. Mol. Evol. 64, 591–600 (2007).
Goddard, M. R., Leigh, J., Roger, A. J. & Pemberton, A. J. Invasion and persistence of a selfish gene in the Cnidaria. PLoS ONE 1, e3 (2006).
Keeling, P. J. et al. The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676 (2005).
Burki, F. et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE 2, e790 (2007).
Gladyshev, E. A., Meselson, M. & Arkhipova, I. R. Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213 (2008).
Acknowledgements
The authors wish to thank numerous colleagues for supplying unpublished data and several of the photos in Figure 2 (see the figure for photo credits). P.J.K.'s research on HGT is supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Tula Foundation, and he is a Fellow of the Canadian Institute for Advanced Research. J.D.P.'s research on HGT is supported by a grant from the National Institutes of Health and by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information S1 (table)
Number of genes putatively acquired from prokaryotesa in heavily sequenced protists (PDF 127 kb)
Related links
Glossary
- Polytomy
-
In a phylogeny, when a branching pattern cannot be resolved, the branches in question can be collapsed to show the absence of a hypothesis for the relationships among the lineages that they represent.
- Endocytosis
-
A general eukaryotic cellular process using the cytoskeleton and endomembrane system to take up material from the environment.
- Ectosymbiont
-
An organism living in a symbiotic association with another, specifically by attachment to the surface of its host.
- Endosymbiont
-
An organism living in a symbiotic association with another, specifically by living inside a host cell.
- Rumen
-
A fermentative compartment of the digestive system in many cellulose-digesting vertebrates, the contents of which are rich in anaerobic protists and prokaryotes.
- Ciliate
-
A lineage of protists (for example, Tetrahymena and Paramecium), predominantly predators, defined by the presence of dimorphic nuclei and large numbers of short flagella (cilia) on the surface. They are members of the Chromalveolates.
- Trypanosomid
-
A lineage of protist flagellates (for example, Trypanosoma, the sleeping-sickness agent), predominantly made up of parasites, and home to many unusual characteristics of genome structure (for example, RNA editing). They are members of the Excavates.
- Diplomonad
-
A lineage of anaerobic or microaerophic protist flagellates (for example, Giardia lamblia), predominantly parasitic and often studied because of their reduced metabolism and mitochondria. They are members of the Excavates.
- Parabasalian
-
A lineage of anaerobic or microaerophic protist flagellates (for example, Trichomonas), predominantly parasitic and often studied owing to their reduced metabolism and their hydrogenosome, a hydrogen-producing mitochondrial relict. They are members of the Excavates.
- Dinoflagellate
-
A lineage of protist flagellates (for example, Alexandrium, a red tide alga) with photosynthetic, heterotrophic and parasitic representatives, which are known for many unusual modifications to genome structure — they are members of the Chromalveolates.
- Oomycete
-
A lineage of protist parasites (for example, Phytophthora, the potato late-blight agent) that are responsible for numerous plant diseases, and were once mistakenly thought to be fungi but are really heterokonts. They are members of the Chromalveolates.
- Osmotrophy
-
Feeding by absorption of nutrients directly from the environment (which can include a host organism in the case of parasites).
- Heterokont
-
A lineage of protist (for example, oomycete parasites and kelps) with photosynthetic, heterotrophic and parasitic representatives, all of which are united by the possession of uniquely dimorphic flagella. They are members of the Chromalveolates.
- Haptophyte
-
A lineage of photosynthetic protist (for example, Emiliania), predominantly marine, some of which form massive marine blooms, and many of which make distinctive calcium carbide scales that have contributed significantly to limestone deposits. They are members of the Chromalveolates.
- Chlorarachniophyte
-
A lineage of photosynthetic protist (for example, Bigelowiella) with amoeboid and flagellate life stages, best known for their retention of a relict nucleus of their green algal plastid endosymbiont, known as a nucleomorph. They are members of the Rhizaria.
- Monophyly
-
In phylogeny, a common ancestor and all its descendants are monophyletic (for example, animals), as opposed to a collection of organisms that does not include their common ancestor, which are polyphyletic (for example, flying animals). Monophyletic is sometimes subdivided into holophyletic (the most recent common ancestor and all things that evolved from it, for example, animals) and paraphyletic (the most recent common ancestor, but not all the things derived from it, for example, reptiles — from which birds evolved).
- Chromalveolates
-
A hypothetical 'supergroup' of protists, including apicomplexa, dinoflagellates, ciliates, heterokonts, haptophytes and cryptomonads, all of which are hypothesized to have diverged from an ancient common ancestor that has acquired a plastid by secondary endosymbiosis with a red alga.
Rights and permissions
About this article
Cite this article
Keeling, P., Palmer, J. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9, 605–618 (2008). https://doi.org/10.1038/nrg2386
Issue Date:
DOI: https://doi.org/10.1038/nrg2386
This article is cited by
-
An expectation–maximization algorithm for estimating proportions of deletions among bacterial populations with application to study antibiotic resistance gene transfer in Enterococcus faecalis
Marine Life Science & Technology (2023)
-
Multiple horizontal transfers of a Helitron transposon associated with a parasitoid wasp
Mobile DNA (2022)
-
A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins
Nature Communications (2022)
-
Cell type-specific intercellular gene transfer in mammalian cells via transient cell entrapment
Cell Discovery (2022)