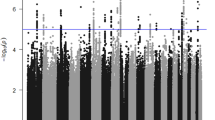
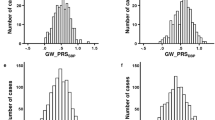
Key Points
-
Arterial pressure is determined by the complex interactions of cardiovascular haemodynamics, kidney function and neural, endocrine and paracrine factors. The function of the kidneys in regulating fluid and electrolyte balance has been shown to be the most important long-term determinant of blood pressure. Essential hypertension represents an imbalance of one or more of these determinants of arterial pressure and is the most common cardiovascular disease, being one of the main risk factors for stroke, heart disease and end-stage kidney disease.
-
Defining the genetic basis of susceptibility to hypertension is challenging because of the complex polygenic nature of arterial blood pressure, which is a quantitative trait that is influenced by multiple variants, gene–gene interactions and environmental factors.
-
Genetic sequence variations within families that are affected by Mendelian forms of hypertension have been identified, but these rare alleles account for less than 1% of human hypertension. However, all of these mutations were found to affect renal tubular electrolyte transport functions, confirming the importance of the kidney in the regulation of blood pressure.
-
Linkage studies using single polymorphic markers that were previously known to participate in a biological pathway of interest (candidate gene markers) have not shown strong linkage with hypertension, and replication between populations has been limited.
-
Genome-scanning linkage studies using microsatellite markers or SNPs have become the method of choice in hypertension research, and have identified many QTLs across the genome. These QTLs have not yet been mapped to the level of individual genetic variants, but attention to precise phenotyping and ecogenetic context is likely to be critically important in this respect.
-
Isolated founder populations show reduced genetic and environmental heterogeneity and provide greater power for QTL mapping because of longer linkage disequilibrium intervals. Studies in such populations have allowed the mapping of QTLs that underlie both arterial pressure per se and intermediate phenotypes that contribute to blood pressure variation.
-
Several candidate-gene association studies for hypertension have been carried out with normotensive (control) and hypertensive (case) subjects. Such approaches are providing fruitful results in other complex diseases. Furthermore, to identify variants across the genome in an unbiased manner, the large numbers of SNPs that have been identified and characterized by efforts such as the HapMap project provide the basis for future genome-wide association studies for hypertension.
-
Rodent models overcome many of the confounding issues that are related to the genetic and environmental heterogeneity that is present in human populations. The rat enables the production of large numbers of informative progeny, invasive measurements and mechanistic studies, which are important not only for phenotyping for the initial ascertainment of QTL, but even more so for narrowing QTL regions.
-
A number of inbred rat strains have been developed that mimic various aspects of human essential hypertension. Comparative mapping to conserved regions in the human genome, and confirmation of linkage across species, seems to be a promising approach for narrowing the regions of interest.
-
Linkage data from rat intercross studies have identified many blood pressure QTLs on a range of chromosomes. These linkage studies confirm the polygenic nature of hypertension and, as found in human linkage studies, dense clusters of blood-pressure-related QTLs are present on a subset of chromosomes.
-
Chromosomal substitution techniques between inbred rats with homogeneous genetic backgrounds provide powerful tools to confirm and narrow down QTL regions in the form of inbred consomic and congenic strains.
-
Microarrays are now being used to identify gene expression differences between normotensive and hypertensive strains. These techniques are beginning to define the molecular, biochemical and physiological pathways that are involved in hypertension and could point to candidate genes. However, to assess causal relationships it is important to carry out serial studies that can properly identify those genes with a relevant temporal response.
-
Genomic regions that are identified as important in regulating blood pressure in rat models of hypertension could provide useful guides in the discovery of functionally related conserved genomic regions in human populations. These model systems will also be essential for determining the functions of newly discovered human candidate genes, through invasive phenotyping and transgenesis.
Abstract
QTL mapping in humans and rats has identified hundreds of blood-pressure-related phenotypes and genomic regions; the next daunting task is gene identification and validation. The development of novel rat model systems that mimic many elements of the human disease, coupled with advances in the genomic and informatic infrastructure for rats, promise to revolutionize the hunt for genes that determine susceptibility to hypertension. Furthermore, methods are evolving that should enable the identification of candidate genes in human populations. Together with the computational reconstruction of regulatory networks, these methods provide opportunities to significantly advance our understanding of the underlying aetiology of hypertension.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pickering, G. W. High Blood Pressure 2nd edn (Grune & Stratton, New York, 1968).
Janeway, T. C. The Clinical Study of Blood Pressure (D. Appleton & Co., New York, 1904).
Schroeder, H. A. Hypertensive Diseases — Causes and Control (Lea & Febiger, Philadelphia, 1953).
Kearney, P. M., et al. Global burden of hypertension: analysis of worldwide data. Lancet 365, 217–223 (2005). Describes how the public-health challenge of hypertension is increasing worldwide, indicating that the prevention, detection, treatment and control of this condition should be a high priority.
Hajjar, I., Kotchen, T. A. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA 290, 199–206 (2003).
Lalouel, J. M. Large-scale search for genes predisposing to essential hypertension. Am. J. Hypertens. 16, 163–166 (2003). Describes the primary objective of the Family Blood Pressure Program initiated by the US National Heart, Lung and Blood Institute, which is to identify the genetic determinants of essential hypertension in humans.
Lee, W. K., Padmanabhan, S. & Dominiczak, A. F. Genetics of hypertension: from experimental models to clinical applications. J. Hum. Hypertens. 14, 631–647 (2000).
Lifton, R. P. & Jeunemaitre, X. Finding genes that cause human hypertension. J. Hypertens. 11, 236–239 (1993).
Cowley, A. W. Jr. Long-term control of blood pressure. Physiol. Rev. 72, 231–300 (1992). This review explores the two primary determinants of long-term regulation of arterial pressure — neural control and volume regulation.
Hall, J. E. The kidney, hypertension, and obesity. Hypertension 41, 625–633 (2003).
Guyton, A. C., Coleman T. G. & Granger, H. J. Circulation: overall regulation. Annu. Rev. Physiol. 34, 13–46 (1972).
Guyton, A. C. Blood pressure control-special role of the kidneys and body fluids. Science 252, 1813–1816 (1991). The first study to show that the kidney dominates long-term blood pressure control through its role in regulating body-fluid volume.
Touyz, R. M. Reactive oxygen species, vascular oxidative stress, and redox signaling in hypertension: what is the clinical significance? Hypertension 44, 248–252 (2004).
Campese, V. M. Salt sensitivity in hypertension. Hypertension 23, 531–550 (1994).
Jones, C. A. Hypertension and renal dysfunction: NHANES III. J. Am. Soc. Nephrol. 14, S71–S75 (2003).
Kotchen, T. A. et al. Glomerular hyperfiltration in hypertensive African Americans. Hypertension 35, 822–826 (2000).
El-Gharbawy, A. H., et al. Predictors of target organ damage in hypertensive blacks and whites. Hypertension 38, 761–766 (2001).
Cooper, R. & Rotimi, C. Hypertension in blacks. Am. J. Hypertens. 10, 804–812 (1997).
Cooper, R. S. et al. An international comparative study of blood pressure in populations of European vs. African descent. BMC Med. 5, e2 (2005).
Jiang, J. et al. Transfer of a salt-resistant renin allele raises blood pressure in Dahl salt-sensitive rats. Hypertension 29, 619–627 (1997).
Yagil, C. et al. Salt susceptibility maps to chromosomes 1 and 17 with sex specificity in the Sabra rat model of hypertension. Hypertension 31, 119–124 (1998).
Cowley, A. W. Jr et al. Genetically defined risk of salt-sensitivity in an intercross of Brown Norway and Dahl S rats. Physiol. Genomics 2, 107–115 (2000).
Provoost, A. P., Shiozawa, M., Van Dokkum, R. P. & Jacob, H. J. Transfer of the Rf-1 region from FHH onto the ACI background increases susceptibility to renal impairment. Physiol. Genomics 8, 123–129 (2002).
Schulz, A., et al. A major gene locus links early onset albuminuria with renal interstitial fibrosis in the MWF rat with polygenetic albuminuria. J. Am. Soc. Nephrol. 14, 3081–3089 (2003).
Hajjar, I. M., Grim, C. E., George, V. & Kotchen, T. A. Impact of diet on blood pressure and age-related changes in blood pressure in the US population: analysis of NHANES III. Arch. Intern. Med. 161, 589–593 (2001).
Luft, F. C. et al. Salt sensitivity and resistance of blood pressure. Age and race as factors in physiological responses. Hypertension 17, 1102–1108 (1991).
Lifton, R. P. Molecular genetics of human blood pressure variation. Science 272, 676–680 (1996).
Mansfield, T. A. et al. Multilocus linkage of familial hyperkalaemia and hypertension, pseudohypoaldosteronism type II, to chromosomes 1q31–42 and 17p11–q21. Nature Genet. 16, 202–205 (1997).
Lifton, R. P., Gharavi, A. G. & Geller, D. S. Molecular mechanisms of human hypertension. Cell 104, 545–556 (2001). This review describes the substantial progress that has been made using molecular genetics to identify the fundamental pathways that underlie the pathogenesis of Mendelian forms of hypertension.
Kahle, K. T., Wilson, F. H. & Lifton, R. P. Regulation of diverse ion transport pathways by WNK4 kinase: a novel molecular switch. Trends Endocrinol. Metab. 16, 98–103 (2005).
Wilson, F. H. et al. Human hypertension caused by mutations in WNK kinases. Science 293, 1107–1112 (2001).
Wilson, F. H. et al. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc. Natl Acad. Sci. USA 100, 680–684 (2003).
Wilson, F. H. et al. A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science 306, 1190–1194 (2004).
Lifton, R. P. et al. A chimaeric 11β-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature 355, 262–265 (1992).
Lifton, R. P. et al. Hereditary hypertension caused by chimaeric gene duplications and ectopic expression of aldosterone synthase. Nature Genet. 2, 66–74 (1992).
Mune, T., Rogerson, F. M., Nikkila, H., Agarwal, A. K. & White, P. C. Human hypertension caused by mutations in the kidney isozyme of 11 β-hydroxysteroid dehydrogenase. Nature Genet. 10, 394–399 (1995).
Geller, D. S. et al. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science 289, 119–123 (2000).
Shimkets, R. A. et al. Liddle's syndrome: heritable human hypertension caused by mutations in the β subunit of the epithelial sodium channel. Cell 79, 407–414 (1994).
Hansson, J. H. et al. A de novo missense mutation of the β subunit of the epithelial sodium channel causes hypertension and Liddle's syndrome identifying a proline-rich segment critical for regulation of channel activity. Proc. Natl Acad. Sci. USA 92, 11495–11499 (1995).
Disse-Nicodeme, S. et al. A new locus on chromosome 12p13.3 for pseudohypoaldosteronism type II, an autosomal dominant form of hypertension. Am. J. Hum. Genet. 67, 302–310 (2000).
Barroso, I. et al. Dominant negative mutations in human PPARγ associated with severe insulin resistance, diabetes mellitus, and hypertension. Nature 402, 880–883 (1999).
Luft, F. C. Present status of genetic mechanisms in hypertension. Med. Clin. North Am. 88, 1–18 (2004).
Gong, M. & Hubner, N. Molecular genetics of human hypertension. Clin. Sci. 110, 315–326 (2006).
Kato, N. & Julier, C. Linkage mapping for hypertension susceptibility genes. Curr. Hypertens. Rep. 1, 15–24 (1999).
Agarwal, A., Williams, G. H., & Fisher, N. D. Genetics of human hypertension. Trends Endocrinol. Metab. 16, 127–133 (2005). A review of candidate-gene studies for human hypertension. Emphasizes that the use of intermediate phenotypes and dense mapping of candidate genes would provide a better approach for identifying genotype–phenotype correlations for human hypertension.
Knight, J., Monroe, P. B., Pembroke, J. C. & Caulfield, M. J. Human chromosome 17 in essential hypertension. Ann. Hum. Gen. 67, 193–206 (2003).
Tikhonoff, V. et al. β-Adducin polymorphisms, blood pressure, and sodium excretion in three European populations. Am. J. Hypertens. 16, 840–846 (2003).
Lander, E. S. & Schork, N. J. Genetic dissection of complex traits. Science 265, 2037–2048 (1994).
Botstein, D. & Risch, N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nature Genet. 33, 228–237 (2003).
Mayeux, R. Mapping the new frontier: complex genetic disorders. J. Clin. Invest. 115, 1404–1407 (2005).
International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005). Describes the HapMap resource, which has characterized more than one million SNPs and can guide the design and analysis of genetic association studies.
Kruglyak, L. Power tools for human genetics. Nature Genet. 37, 1299–1300 (2005).
Thorisson, G. A., Smith, A. V., Krishnan, L. & Stein, L. D. The International HapMap Project web site. Genome Res. 15, 1592–1593 (2005).
Kuznetsova T. et al. Sodium excretion as a modulator of genetic associations with cardiovascular phenotypes in the European Project on Genes in Hypertension. J. Hypertens. 24, 235–242 (2006).
Mattson, D. L., Meister, C. J., Marcelle, M. L. Dietary protein source determines the degree of hypertension and renal disease in the Dahl salt-sensitive rat. Hypertension 45, 736–741 (2005).
Newman, D. L. et al. Major loci influencing serum triglyceride levels on 2q14 and 9p21 localized by homozygosity-by-descent mapping in a large Hutterite pedigree. Hum. Mol. Genet. 12, 137–144 (2003).
O'Brien, E., Kerber, R. A., Jorde, L. B. & Rogers, A. R. Founder effect: assessment of variation in genetic contributions among founders. Hum. Biol. 66, 185–204 (1994).
Jeunemaître, X. et al. Molecular basis of human hypertension: role of angiotensinogen. Cell 71, 169–180 (1992).
Hata, A. et al. Angiotensinogen as a risk factor for essential hypertension in Japan. J. Clin. Invest. 3, 1285–1287 (1994).
Wu X. et al. An association study of angiotensinogen polymorphisms with serum level and hypertension in an African-American population. J. Hypertens. 10, 1847–1852 (2003).
Hamet, P. et al. Quantitative founder-effect analysis of French Canadian families identifies specific loci contributing to metabolic phenotypes of hypertension. Am J. Hum. Genet. 76, 815–832 (2005). Describes the assessment of a founder effect, whereby traits that are determined within population subsets are measurably and quantitatively transmitted through a generational lineage, in a French Canadian population with essential hypertension. Numerous QTLs were found, with prominent clusters located on two chromosomes.
Bouchard, G., Roy, R., Casgrain, B. & Hubert, M. [Population files and database management: the BALSAC database and the INGRES/INGRID system]. Hist. Mes. 4, 39–57 (1989) (in French).
Kotchen, T. A. et al. Identification of hypertension-related QTLs in African American sib pairs. Hypertension 40, 634–639 (2002).
Langefeld, C. D. et al. Linkage of the metabolic syndrome to 1q23–q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes 53, 1170–1174 (2004).
Hanson, R. L. et al. Autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am. J. Hum. Genet. 63, 1130–1138 (1998).
Binder, A. Identification of genes for a complex trait: examples from hypertension. Curr. Pharm. Biotechnol. 7, 1–13 (2006).
Hirschhorn, J. N. & Daly, M. J. Genome-wide association studies for common diseases and complex traits. Nature Rev. Genet. 6, 95–108 (2005).
Todd, J. A. Statistical false positive or true disease pathway? Nature Genet. 38, 731–733 (2006).
Hinds, D. A. et al. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 (2005).
Gretarsdottir, S. et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nature Genet. 35, 131–138 (2003).
Kuhlenbaumer, G. et al. Evaluation of single nucleotide polymorphisms in the phosphodiesterase 4D gene (PDE4D) and their association with ischemic stroke in a large German cohort. J. Neurol. Neurosurg. Psychiatry 77, 521–524 (2006).
Marguiles, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 1728–1732 (2005).
Shendure, J. et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309, 1728–1732 (2005).
Stoll, M. et al. New target regions for human hypertension via comparative genomics. Genome Res. 10, 473–482 (2000). This study presents a comparative genomic map for candidate hypertension loci in humans on the basis of QTLs that show genetic homology between rats, mice and humans.
Dahl, L. K., Heine, M. & Tassinari, L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature 5, 480–482 (1962).
Rapp, J. P. Dahl salt-susceptible and salt-resistant rats. Hypertension 4, 753–763 (1982).
Jacob, H. J. et al. Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell 67, 213–224 (1991).
Koike, G. et al. Mapping of the rat Sm22 gene to chromosome 8q24: a candidate for high blood pressure and cardiac hypertrophy. Mamm. Genome 6, 216–218 (1995).
Brown, D. M., Provoost, A. P., Daly, M. J., Lander, E. S. & Jacob, H. J. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nature Genet. 12, 44–51 (1996).
Harris, E. L. et al. Strain-specific deletions in exon 10 of rat K-kininogen and T-kininogen genes allow mapping of both genes to rat chromosome 11. Mamm. Genome 8, 791–792 (1997).
Innes, B. A., McLaughlin, M., Kapuscinski, M. K. & Jacob, H. J. Independent genetic susceptibility of cardiac hypertrophy in inherited hypertension. Hypertension 31, 741–746 (1998).
Rapp, J. P. Genetic analysis of inherited hypertension in the rat. Phys. Rev. 80, 135–172 (2000). A review of the genetic dissection of hypertension using selectively bred strains of rats with divergent blood pressures. The theoretical basis and methodologies used are covered, including QTL analyses and the use of congenic strains.
Gibbs, R. A. et al. Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521 (2004). This paper reports the first high-quality draft of the genome of the Brown Norway rat, covering over 90% of the genome sequence.
Michalkiewicz, M et al. Transgenic rescue demonstrates involvement of the Ian5 gene in T cell development in the rat. Physiol. Genomics. 9, 228–232 (2004).
Cowley, A. W. Jr et al. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension 37, 456–461 (2001).
Stoll, M. et al. A genomic-systems biology map for cardiovascular function. Science 294, 1723–1726 (2001).
Kato, N. et al. Genome-wide searches for blood pressure quantitative trait loci in the stroke-prone spontaneously hypertensive rat of a Japanese colony. J. Hypertens. 21, 295–303 (2003).
Yagil, C., Hubner, N., Kreutz, R., Ganten, D. & Yagil, Y. Congenic strains confirm the presence of salt-sensitivity QTLs on chromosome 1 in the Sabra rat model of hypertension. Physiol. Genomics 12, 85–95 (2003).
Moreno, C. et al. Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol. Genomics 15, 243–257 (2003).
Glazier, A. M., Nadeau, J. H., Aitman, J. T. Finding genes that underlie complex traits. Science 298, 2345–2349 (2002).
Yagil, Y. & Yagil, C. Problems with linkage analysis and QTL detection in hypertension. J. Hypertens. 21, 247–249 (2003).
Rapp, J. P., Wang, S. M. & Dene, H. Effect of genetic background on cosegregation of renin alleles and blood pressure in Dahl rats. Am. J. Hypertens. 3, 391–396 (1990).
Cicila, G. T. et al. Cosegregation of the endothelin-3 locus with blood pressure and relative heart weight in inbred Dahl rats. J. Hypertens. 12, 643–651 (1994).
Abiola, O. et al. The Complex Trait Consortium. The nature and identification of quantitative trait loci: a community's view. Nature Rev. Genet. 4, 911–916 (2003). A proposal for the standardization of the approaches and statistical analyses that are required for the identification of genetic loci that determine quantitative traits.
Pravenec, M. et al. Transgenic rescue of defective Cd36 ameliorates insulin resistance in spontaneously hypertensive rats. Nature Genet. 27, 156–158 (2001).
Pfeifer, A., Ikawa, M., Dayn, Y. & Verma, I. M. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl Acad. Sci. USA 99, 2140–2145 (2002).
Michalkiewicz, M. et al. Efficient transgenic rat production by a lentiviral vector. FASEB J. 20, 407 (2006).
Nadeau, J. H., Singer, J., Matin, A. & Lander, E. Analysing complex genetic traits with chromosome substitution strains. Nature Genet. 24, 221–225 (2000).
Singer, J. B. et al. Genetic dissection of complex traits with chromosome substitution strains of mice. Science 304, 445–448 (2004).
Cowley, A. W. Jr, Liang, M., Roman, R. J., Greene, A. S. & Jacob, H. J. Consomic rat model systems for physiological genomics. Acta Physiol. Scand. 181, 585–592 (2004). A review of the use of consomic rat strains, a resource that can extend our understanding of genes and their roles in complex function and disease.
Cowley, A. W. Jr, Roman, R. J. & Jacob, H. J. Application of chromosomal substitution techniques in gene function discovery. J. Physiol. 554, 46–55 (2004).
Lee, S. J. et al. Use of a panel of congenic strains to evaluate differentially expressed genes as candidate genes for blood pressure quantitative trait loci. Hypertens. Res. 26, 75–87 (2003).
Steen, R. G. et al. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9, AP1–AP8 (1999).
Charron, S. et al. Epistasis, not numbers, regulates functions of clustered Dahl rat quantitative trait loci applicable to human hypertension. Hypertension 46, 1300–1308 (2005).
Moreno, C. et al. Existence of multiple blood pressure loci on rat chromosome 13 of the Dahl S hypertensive rat. FASEB J. 20, A407 (2006).
Kwitek-Black, A. E., Jacob, H. J. The use of designer rats in the genetic dissection of hypertension. Curr. Hypertens. Rep. 3, 12–18 (2001). Describes the use of consomic and congenic strains, which share phenotypic and genotypic characteristics with humans and are powerful platforms for functional studies.
Hubner, N, Yagil, C & Yagil, Y. Novel integrative approaches to the identification of candidate genes in hypertension. Hypertension 47, 1–5 (2006).
Hubner, N. et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nature Genetics 37, 243–253 (2005).
Liang, M. et al. Insights into Dahl salt-sensitive hypertension revealed by temporal patterns of renal medullary gene expression. Physiol. Genomics 12, 229–237 (2003).
Liang, M., Cowley, A. W. & Greene, A. S. High throughput gene expression profiling: a molecular apporach to integrative physiology. J. Physiol. 554, 22–30 (2004).
Storey, J. D., Xiao, W., Leek, J. T., Tompkins, R. G. & Davis, R. W. Significance analysis of time course microarray experiments. Proc. Natl Acad. Sci. USA 102, 12837–12842 (2005).
Kaneko, Y., Herrera, V. L., Didishvili, T. & Ruiz-Opazo, N. Sex-specific effects of dual ET-1/ANG II receptor (Dear) variants in Dahl salt-sensitive/resistant hypertension rat model. Physiol. Genomics 20, 157–164 (2005).
Weiss, L. A., Pan, L., Abney, M. & Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nature Genet. 38, 218–222 (2006). This paper provides evidence that sex-specific heritability is likely to have important implications for mapping complex traits in genome-wide linkage studies.
Ober, C., Pan, L., Phillips, N., Parry, R., & Kurina, L. M. Sex-specific genetic architecture of asthma-associated quantitative trait loci in a founder population. Curr. Allergy Asthma Rep. 6, 241–246 (2006).
Carrel, L. & Willard, H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005).
Peter, I. et al. Association of estrogen receptor β gene polymorphisms with left ventricular mass and wall thickness in women. Am. J. Hypertens. 18, 1388–1395 (2005).
Rinn, J. L. & Snyder, M. Sexual dimorphism in mammalian gene expression. Trends Genet. 21, 298–305 (2005).
Clough, R. W., Aravich, P. F. & Sladek, C. D. Monosodium glutamate neurotoxicity: a sex-specific impairment of blood pressure but not vasopressin in developing rats. Brain Res. Bull. 17, 51–58 (1986).
Krajnak, K., Kashon, M. L., Rosewell, K. L. & Wise, P. M. Sex differences in the daily rhythm of vasoactive intestinal polypeptide but not arginine vasopressin messenger ribonucleic acid in the suprachiasmatic nuclei. Endocrinology 139, 4189–4196 (1998).
Ibrahim, J., McGee, A., Graham, D., McGrath, J. C. & Dominiczak, A. F. Sex-specific differences in cerebral arterial myogenic tone in hypertensive and normotensive rats. Am. J. Physiol. Heart Circ. Physiol. 290, H1081–H1089 (2006).
Acknowledgements
The ideas represented in this overview of the genetics of hypertension have evolved over more than a decade of collaborations with a research team of dedicated and highly interactive geneticists, physiologists and computational biologists working at the Medical College of Wisconsin (MCW). These include H. Jacob, R. Roman, A. Greene, A. Kwitek, M. Olivier, M. Michalkiewicz, M. Liang, J. Lombard, D. Mattson, H. Forster, C. Moreno-Quinn, P. Tonellato, D. Beard and M. Kaldunski. The clinical collaborators responsible for the human hypertension studies in our group are T. Kotchen and U. Broeckel at MCW and P. Hamet at the University of Montreal. We acknowledge the National Heart Lung and Blood Institute and the National Human Genome Institute for support of this research.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Related links
Glossary
- Compliance
-
The ratio of the increase in the intraluminal volume relative to an increase in pressure within a blood vessel.
- Glomerular filtration
-
The production of plasma ultrafiltrate by glomeruli
- Linkage studies
-
Studies that are designed to identify the co-segregation of marker alleles and disease within pedigrees.
- Renin–angiotensin–aldosterone system
-
The renin–angiotensin system (RAS) and aldosterone hormone systems have a crucial role in sodium and blood-volume homeostasis, and in the long-term regulation of arterial pressure.
- Pleiotropy
-
Describes situations in which one gene contributes to many phenotypic expressions.
- Penetrance
-
The proportion of individuals with a specific genotype who manifest that genotype at the phenotypic level.
- Founder population
-
The small founding population of a new location that subsequently grows and populates the region; the offspring of these founders can be used in genetic studies to reduce variation due to heterogeneity.
- Linkage disequilibrium
-
The non-random associations of alleles. For example, if the proportion of double homozygotes is greater than predicted from normal Mendelian segregation, then there is linkage disequilibrium between the two alleles. This can arise from epistatic selection, and might indicate a functional interaction between loci that is associated with a phenotype of interest.
- Metabolic syndrome
-
A cluster of conditions that often occur together, including obesity, high blood glucose, high blood pressure and high triglyceride levels, which can lead to cardiovascular disease.
- Gene association studies
-
Population-based genetic studies that examine whether an allele or marker segregates with a phenotype or disease at a significantly higher rate than predicted by chance alone. This is ascertained by genotyping variants in both affected and unaffected individuals.
- Genome-wide association studies
-
The scanning of genomes with genetic markers at regular intervals to identify chromosomal regions with elevated levels of marker similarity in normotensive compared with hypertensive subjects.
- Consomic strain
-
An animal strain that is produced by transferring a single, full-length chromosome from one inbred strain (the donor) into the genetic background of a host strain (the recipient) by repeated backcrossing.
- Congenic strain
-
A congenic strain is produced by transferring a part of a chromosome from one inbred strain into the genetic background of a host strain.
- Marker-assisted selection
-
The use of genetic markers for the selection of a linked characteristic, trait or disease-associated gene.
- Dimension reduction
-
A computational technique that is widely used in informatics science for the mining and visualization of large-scale data sets.
Rights and permissions
About this article
Cite this article
Cowley, A. The genetic dissection of essential hypertension. Nat Rev Genet 7, 829–840 (2006). https://doi.org/10.1038/nrg1967
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg1967
This article is cited by
-
Nestin expression in intact and hypertrophic myocardium of spontaneously hypertensive rats during aging
Journal of Muscle Research and Cell Motility (2023)
-
Natural selection and local adaptation of blood pressure regulation and their perspectives on precision medicine in hypertension
Hereditas (2019)
-
Genetics of Resistant Hypertension: the Missing Heritability and Opportunities
Current Hypertension Reports (2018)
-
Unravelling the Lesser Known Facets of Angiotensin II Type 1 Receptor
Current Hypertension Reports (2017)
-
Characteristics of Long Non-coding RNAs in the Brown Norway Rat and Alterations in the Dahl Salt-Sensitive Rat
Scientific Reports (2014)