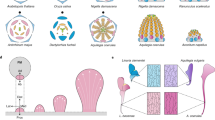
Abstract
The Mostly Male theory is the first to use evidence from gene phylogenies, genetics, modern plant morphology and fossils to explain the evolutionary origin of flowers. It proposes that flower organization derives more from the male structures of ancestral gymnosperms than from female structures. The theory arose from a hypothesis-based study. Such studies are the most likely to generate testable evolutionary scenarios, which should be the ultimate goal of evo-devo.
This is a preview of subscription content, access via your institution
Access options





Similar content being viewed by others
References
Darwin, F. & Seward, A. C. (eds) More Letters of Charles Darwin: A Record of his Work in a Series of Hitherto Unpublished Letters Vol. 2 20–21 (John Murray, London, 1903).
Stebbins, G. L. Flowering Plants: Evolution Above the Species Level (Harvard Univ. Press, Cambridge, Massachusetts, 1974).
Doyle, J. A. Origin of angiosperms. Annu. Rev. Ecol. Syst. 9, 365–392 (1978).
Taylor, D. W. & Hickey, L. J. (eds) Flowering Plant Origin, Evolution and Phylogeny (Chapman and Hall, New York, 1996).
Crane, P. R. Phylogenetic analysis of seed plants and the origin of angiosperms. Ann. Missouri Bot. Gard. 72, 716–793 (1985).
Friis, E. M. et al. The Origin of Angiosperms and their Biological Consequences (Cambridge Univ. Press, Cambridge, UK, 1987).
Frohlich, M. W. & Parker, D. S. The Mostly Male theory of flower evolutionary origins: from genes to fossils. Syst. Bot. 25, 155–170 (2000).
Frohlich, M. W. in Beyond Heterochrony: The Evolution of Development (ed. Zelditch, M. L.) 59–106 (John Wiley, New York, 2001).
Frohlich, M. W. in Developmental Genetics and Plant Evolution: Systematics Association Special Volume Series 65 (eds Cronk, Q. C. B. et al.) 85–108 (Taylor and Francis, London, 2002).
Doyle, J. A. Seed plant phylogeny and the relationships of Gnetales. Int. J. Plant Sci. 157, 3–39 (1996).
Frohlich M. W. & Meyerowitz, E. M. The search for flower homeotic gene homologs in basal angiosperms and gnetales: a potential new source of data on the evolutionary origin of flowers. Int. J. Plant Sci. 158, 131–142 (1997).
Baum, D. A. The evolution of plant development. Curr. Opin. Plant Biol. 1, 79–86 (1998).
Lohmann, J. U. & Weigel, D. Building beauty: the genetic control of floral patterning. Dev. Cell 2, 135–142 (2002).
Donoghue, M. J. & Doyle, J. A. Seed plant phylogeny: demise of the anthophyte hypothesis? Curr. Biol. 10, 106–109 (2000).
Frohlich, M. W. & Estabrook, G. F. Wilkinson support calculated with exact probabilities: an example using Floricaula/LEAFY amino acid sequences that compares three hypotheses involving gene gain/loss in seed plants. Mol. Biol. Evol. 17, 1914–1925 (2000).
Mouradov, A. et al. NEEDLY, a Pinus radiata ortholog of FLORICAULA/LEAFY genes, expressed in both reproductive and vegetative meristems. Proc. Natl Acad. Sci. USA 95, 6537–6542 (1998).
Mellerowicz, E. J. et al. PRFLL a Pinus radiata homologue of FLORICAULA and LEAFY is expressed in buds containing vegetative shoot and undifferentiated male cone primordia. Planta 206, 619–629 (1998).
Colombo, L. et al. The Petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868 (1995).
Decraene, L. P. R. & Smets, E. F. Notes on the evolution of androecial organisation in the Magnoliophytina (angiosperms). Bot. Acta 111, 77–86 (1998).
Hufford, L. The morphology and evolution of male reproductive structures of Gnetales Int. J. Plant Sci. 157, 95–112 (1996).
Endress, P. K. Structure and function of female and bisexual organ complexes in Gnetales. Int. J. Plant Sci. 157, 113–125 (1996).
Yao, X. et al. The Corystosperm pollen organ Pteruchus from the Triassic of Antarctica. Amer. J. Bot. 82, 535–546 (1995).
Klavins, S. D. et al. Anatomy of Umkomasia (Corystospermales) from the Triassic of Antarctica. Amer. J. Bot. 89, 664–676 (2002).
Shindo, S. et al. Characterization of a FLORICAULA/LEAFY homologue of Gnetum parvifolium and its implications for the evolution of reproductive organs in seed plants. Int. J. Plant Sci. 162, 1199–1209 (2001).
Long, J. A. et al. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996).
Brown, R. C. & Mogensen, H. L. Late ovule and early embryo development in Quercus gambelii. Amer. J. Bot. 59, 311–316 (1972).
Bowman, J. L. et al. Establishment of polarity in angiosperm lateral organs. Trends Genet. 18, 134–141 (2002).
Svoma, E. Seed development and function in Artabotrys hexapetalus (Annonaceae). Plant Syst. Evol. 207, 205–223 (1997).
Taylor, T. N. et al. Permineralized seed fern cupules from the Triassic of Antarctica — implications for cupule and carpel evolution. Amer. J. Bot. 81, 666–677 (1994).
Griffith, M. E. et al. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 126, 5635–5644 (1999).
Nelson, J. M. et al. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development 129, 4581–4589 (2002).
Groß-Hardt, R. et al. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138 (2002).
Winter, K. U. et al. Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19, 587–596 (2002).
Becker, A. et al. A novel MADS-box gene subfamily with a sister-group relationship to class B floral homeotic genes. Mol. Genet. Genomics 266, 942–950 (2002).
Nesi, N. et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14, 2463–2479 (2002).
Bowe, L. M. et al. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales' closest relatives are conifers. Proc. Natl Acad. Sci. USA 97, 4092–4097 (2000).
Chaw, S. M. et al. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proc. Natl Acad. Sci. USA 97, 4086–4091 (2000).
Qiu, Y. -L. et al. Phylogeny of basal angiosperms: analyses of five genes from three genomes. Int. J. Plant Sci. 161, 3–27 (2000).
Mathews, S. & Donoghue, M. J. Basal angiosperm phylogeny inferred from duplicate phytochromes A and C. Int. J. Plant Sci. 161, 41–55 (2000).
Soltis, D. E. et al. Missing links: the genetic architecture of flower and floral diversification. Trends Plant Sci. 7, 22–31 (2002).
Lanfranchi, G. et al. Identification of 4370 expressed sequence tags from a 3′-end-specific cDNA library of human skeletal muscle by DNA sequencing and filter hybridization. Genome Res. 6, 35–42 (1996).
Albert, V. A. et al. Pleiotropy, redundancy and the evolution of flowers. Trends Plant Sci. 7, 297–301 (2002).
Theissen, G. et al. in Developmental Genetics and Plant Evolution: Systematics Association Special Volume Series 65 (eds Cronk, Q. C. B. et al.) 173–206 (Taylor and Francis, London, 2002).
Lamark, J. B. Zoological Philosophy (1809) 122 (Univ. Chicago Press, Chicago, 1984) (Translated by H. Elliot).
Mayr, E. The Growth of Biological Thought (Belknap, Cambridge, Massachusetts, 1982).
Frohlich, M. W. MADS about Gnetales. Proc. Natl Acad. Sci. USA 96, 8811–8813 (1999).
Gould, S. J. Sociobiology: the art of storytelling. New Sci. 80, 530–533 (1978).
Gould, S. J. & Lewontin, R. C. The Spandrels of San-Marco and the Panglossian paradigm — a critique of the adaptationist program. Proc. Royal Soc. London B 205, 581–598 (1979).
Iltis, H. H. From teosinte to maize: the catastrophic sexual transmutation. Science 222, 886–894 (1983).
Iltis, H. H. Homeotic sexual translocations and the origin of maize (Zea mays, Poaceae): a new look at an old problem. Econ. Bot. 54, 7–42 (2000).
Lauter, N. & Doebley, J. Genetic variation for phenotypically invariant traits detected in teosinte: implications for the evolution of novel forms. Genetics 160, 333–342 (2002).
Gibson, G. Developmental evolution: going beyond the 'just so'. Curr. Biol. 9, 942–945 (1999).
Acknowledgements
I would like to thank E. Meyerowitz, in whose laboratory this work began, J. Trager of Huntington Gardens, San Marino, California, and L. Song of the University of California, Fullerton, for Welwitschia materials, and two anonymous reviewers. This work is supported by a National Science Foundation grant.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
MaizeGDB
TAIR
FURTHER INFORMATION
Glossary
- CARPEL
-
A leaf homologue that encloses the ovules and seeds in angiosperms and develops into the fruit.
- CHAROPHYTES
-
The group of green algae that are most closely related to land plants.
- CLADISTICS
-
The approach for analysing organismal or gene diversity that uses explicit characters and character states, which are mapped onto branching diagrams that explicitly mirror possible phylogenetic relationships.
- CLADOGRAM
-
A branching diagram that represents organismal or gene phylogeny.
- EVOLUTIONARY SYNTHESIS
-
The movement to deepen the understanding of Darwinian evolution by incorporating knowledge from different fields, including genetics, population genetics, ecology and so on.
- GYMNOSPERMS
-
Plants that make ovules and seeds but lack flowers. There are four extant groups: conifers, cycads, Ginkgo and Gnetales.
- HETEROTOPY
-
An evolutionary process in which a particular structure of an organism forms in new location.
- LONG BRANCH
-
An internode in a phylogenetic tree in which large amounts of character-state change have occured.
- MAD-BOX GENES
-
Eukaryotic genes that have the MADS-box DNA-binding domain.
- MONOPHYLETIC
-
A group of organisms or genes descended from a common ancestor that is a member of the group, which includes all descendents of that common ancestor.
- ORTHOLOGUES
-
Two or more genes (or gene families) that became distinct lineages as the result of a speciation event.
- OUTGROUP
-
A group of organisms or genes that is outside the monophyletic group under consideration.
- PARALOGUES
-
Two or more genes (or gene families) that became distinct lineages owing to a gene-duplication event in an organism.
- PERMINERALIZED
-
A plant fossil resulting from mineral deposition in the cells and tissues of the specimen, which often preserves the cell walls and tissue structure.
- PHYLLARIES
-
Modified leaves that surround the dense flower clusters (heads) of plants in the sunflower family.
- SEPAL
-
The outer-most group of organs in a flower.
Rights and permissions
About this article
Cite this article
Frohlich, M. An evolutionary scenario for the origin of flowers. Nat Rev Genet 4, 559–566 (2003). https://doi.org/10.1038/nrg1114
Issue Date:
DOI: https://doi.org/10.1038/nrg1114
This article is cited by
-
Reciprocal expression of MADS-box genes and DNA methylation reconfiguration initiate bisexual cones in spruce
Communications Biology (2024)
-
A symmetry gene restores femaleness
Nature Plants (2022)
-
Case not closed: the mystery of the origin of the carpel
EvoDevo (2021)
-
A new hybrid model based on relevance vector machine with flower pollination algorithm for phycocyanin pigment concentration estimation
Environmental Science and Pollution Research (2021)
-
Flower pollination algorithm: a comprehensive review
Artificial Intelligence Review (2019)