Abstract
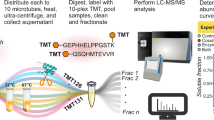
To link cleaved substrates in complex systems with a specific protease, the protease active site specificity is required. Proteomic identification of cleavage sites (PICS) simultaneously determines both the prime- and non-prime-side specificities of individual proteases through identification of hundreds of individual cleavage sequences from biologically relevant, proteome-derived peptide libraries. PICS also identifies subsite cooperativity. To generate PICS peptide libraries, cellular proteomes are digested with a specific protease such as trypsin. Following protease inactivation, primary amines are protected. After incubation with a test protease, each prime-side cleavage fragment has a free newly formed N-terminus, which is biotinylated for affinity isolation and identification by liquid chromatography–tandem mass spectrometry. The corresponding non-prime sequences are derived bioinformatically. The step-by-step protocol also presents a web service for PICS data analysis, as well as introducing and validating PICS peptide libraries made from Escherichia coli.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schechter, I. & Berger, A. On the active site of proteases. 3. Mapping the active site of papain; specific peptide inhibitors of papain. Biochem. Biophys. Res. Commun. 32, 898–902 (1968).
Schilling, O. & Overall, C.M. Proteomic discovery of protease substrates. Curr. Opin. Chem. Biol. 11, 36–45 (2007).
Schilling, O. & Overall, C.M. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 (2008).
Auf dem Keller, U. & Schilling, O. Proteomic techniques and activity-based probes for the system-wide study of proteolysis. Biochimie 92, 1705–1714 (2010).
Matthews, D.J. & Wells, J.A. Substrate phage: selection of protease substrates by monovalent phage display. Science 260, 1113–1117 (1993).
Boulware, K.T. & Daugherty, P.S. Protease specificity determination by using cellular libraries of peptide substrates (CLiPS). Proc. Natl. Acad. Sci. USA 103, 7583–7588 (2006).
Turk, B.E., Huang, L.L., Piro, E.T. & Cantley, L.C. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 19, 661–667 (2001).
Kleifeld, O. et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 (2010).
Sukuru, S.C.K. et al. A lead discovery strategy driven by a comprehensive analysis of proteases in the peptide substrate space. Protein Sci. 19, 2096–2109 (2010).
Schilling, O. & Overall, C.M. Protease subsite profiling with proteome-derived peptide libraries (PICS). Nat. Protoc. doi: 10.1038/nprot.2008.88 (2008).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Craig, R. & Beavis, R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Pedrioli, P.G.A. Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol. Biol. 604, 213–238 (2010).
Colaert, N., Helsens, K., Martens, L., Vandekerckhove, J. & Gevaert, K. Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 (2009).
Acknowledgements
We thank B. Mayer for technical assistance in establishing the simplified PICS library generation workflow. O.S. acknowledges support from the Deutsche Forschungsgemeinschaft (DFG, grants SCHI 871/1-1 and 871/2-1) and the Michael Smith Foundation for Health Research (MSFHR). P.F.H. is supported by Postdoctoral Fellowships of the German Academic Exchange Service (DAAD) and the MSFHR. O.B. acknowledges support from the Swiss National Foundation of Sciences (SNF) and the Canadian Institutes of Health Research (CIHR). C.M.O. is supported by a Canada Research Chair in Metalloproteinase Proteomics and Systems Biology. This work was supported by a grant from the CIHR and from a program project grant in Breast Cancer Metastases from the Canadian Breast Cancer Research Alliance (CBCRA) with funds from the Canadian Breast Cancer Foundation and the Cancer Research Society, as well as by an Infrastructure Grant from MSHFR.
Author information
Authors and Affiliations
Contributions
O.S. and C.M.O. designed and developed the PICS procedure. O.S. and U.a.d.K. developed the web-based analysis platform, P.F.H. established bacterial libraries and O.S., P.F.H. and O.B. optimized the protocol. O.S. drafted the manuscript and O.S. and P.F.H. designed the figures. All authors edited and approved the manuscript.
Corresponding author
Rights and permissions
About this article
Cite this article
Schilling, O., Huesgen, P., Barré, O. et al. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat Protoc 6, 111–120 (2011). https://doi.org/10.1038/nprot.2010.178
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.178
This article is cited by
-
Mouse skin peptidomic analysis of the hemorrhage induced by a snake venom metalloprotease
Amino Acids (2023)
-
Identification and characterization of the proteolytic flagellin from the common freshwater bacterium Hylemonella gracilis
Scientific Reports (2020)
-
Block-based characterization of protease specificity from substrate sequence profile
BMC Bioinformatics (2017)
-
Discovery of a proteolytic flagellin family in diverse bacterial phyla that assembles enzymatically active flagella
Nature Communications (2017)
-
Mass spectrometry-based determination of Kallikrein-related peptidase 7 (KLK7) cleavage preferences and subsite dependency
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.