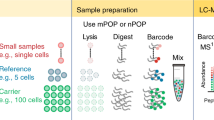
Abstract
'Reverse-phase' protein lysate microarray (RPA) assays use micro-scale, cell lysate dot blots that are printed to a substrate, followed by quantitative immunochemical protein detection, known to be particularly effective across many samples. Large-scale sample collection is a labor-intensive and time-consuming process; the information yielded from RPA assays, however, provides unique opportunities to experimentally interpret theoretical protein networks quantitatively. When specific antibodies are used, RPA can generate 1,000 times more data points using 10,000 times less sample volume than an ordinary western blot, enabling researchers to monitor quantitative proteomic responses for various time-scale and input-dose gradients simultaneously. Hence, the RPA system can be an excellent method for experimental validation of theoretical protein network models. Besides the initial screening of primary antibodies, collection of several hundreds of sample lysates from 1- to 8-h periods can be completed in ∼10 d; subsequent RPA printing and signal detection steps require an additional 2–3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Paweletz, C.P. et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 (2001).
Rudelius, M. et al. Constitutive activation of Akt contributes to the pathogenesis and survival of mantle cell lymphoma. Blood 108, 1668–1676 (2006).
Tibes, R. et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5, 2512–2521 (2006).
Wulfkuhle, J.D. et al. Signal pathway profiling of ovarian cancer from human tissue specimens using reverse-phase protein microarrays. Proteomics 3, 2085–2090 (2003).
Ramalingam, S. et al. Quantitative assessment of the p53-Mdm2 feedback loop using protein lysate microarrays. Cancer Res. 67, 6247–6252 (2007).
Nishizuka, S. et al. Quantitative protein network monitoring in response to DNA damage. J. Proteome Res. 7, 803–808 (2008).
Nishizuka, S. & Spurrier, B. Experimental validation for quantitative protein network models. Curr. Opin. Biotechnol. 19, 41–49 (2008).
Utz, P.J. Protein arrays for studying blood cells and their secreted products. Immunol. Rev. 204, 264–282 (2005).
Liotta, L. & Petricoin, E. Molecular profiling of human cancer. Nat. Rev. Genet. 1, 48–56 (2000).
Chan, S.M. et al. Protein microarrays for multiplex analysis of signal transduction pathways. Nat. Med. 10, 1390–1396 (2004).
Winters, M.E. et al. Supra-additive growth inhibition by a celecoxib analogue and carboxyamido-triazole is primarily mediated through apoptosis. Cancer Res. 65, 3853–3860 (2005).
Sevecka, M. & MacBeath, G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods 3, 825–831 (2006).
Spurrier, B. et al. Antibody screening database for protein kinetic modeling. Proteomics 7, 3259–3263 (2007).
Nishizuka, S. et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl Acad. Sci. USA 100, 14229–14234 (2003).
Nishizuka, S. et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 63, 5243–5250 (2003).
Sasagawa, S., Ozaki, Y., Fujita, K. & Kuroda, S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 7, 365–373 (2005).
Lequin, R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 51, 2415–2418 (2005).
Anderson, L. & Seilhamer, J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18, 533–537 (1997).
Nishizuka, S. Profiling cancer stem cells using protein array technology. Eur. J. Cancer 42, 1273–1282 (2006).
Major, S.M. et al. AbMiner: a bioinformatic resource on available monoclonal antibodies and corresponding gene identifiers for genomic, proteomic, and immunologic studies. BMC Bioinformatics 7, 192 (2006).
Nishizuka, S., Washburn, N.R. & Munson, P.J. Evaluation method of ordinary flatbed scanners for quantitative density analysis. Biotechniques 40, 442, 444, 446 passim (2006).
Carlisle, A.J. et al. Development of a prostate cDNA microarray and statistical gene expression analysis package. Mol. Carcinog. 28, 12–22 (2000).
Ramaswamy, A. et al. Application of protein lysate microarrays to molecular marker verification and quantification. Proteome Sci. 3, 9 (2005).
Calvert, V.S. et al. Development of multiplexed protein profiling and detection using near infrared detection of reverse-phase protein microarrays. Clin. Proteomics J. 1, 81–89 (2004).
Acknowledgements
We thank Drs. John Austin, Toni Holway and Peter Honkanen of Auslon BioSystems for excellent protein microarray technical support, and Dr. Lynn Young for customizing image processing programs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Spurrier, B., Ramalingam, S. & Nishizuka, S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat Protoc 3, 1796–1808 (2008). https://doi.org/10.1038/nprot.2008.179
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.179
This article is cited by
-
Reversed-phase allergen microarrays on optical discs for multiplexed diagnostics of food allergies
Microchimica Acta (2023)
-
A reverse phase protein array based phospho-antibody characterization approach and its applicability for clinical derived tissue specimens
Scientific Reports (2022)
-
Oncoproteomics: insight into current proteomic technologies in cancer biomarker discovery and treatment
Journal of Proteins and Proteomics (2022)
-
Myopathy associated LDB3 mutation causes Z-disc disassembly and protein aggregation through PKCα and TSC2-mTOR downregulation
Communications Biology (2021)
-
The Translational Status of Cancer Liquid Biopsies
Regenerative Engineering and Translational Medicine (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.