Abstract
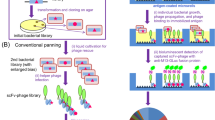
Phage-display of antibody libraries is a powerful tool to select antibodies for specific epitopes. We describe a strategy for selecting highly specific scFv-clones that discriminate between various conformational states of cell surface receptors. This approach adapts the M13 pIII phage-display technology toward a cell suspension-based strategy, which allows panning against complex, multimeric, fully functional cell membrane epitopes without alteration of structure due to purification or immobilization. As the functional properties are preserved, phage can be specifically depleted or selected for neo-epitopes exposed after physiological alterations of the targeted molecules. This subtractive strategy allows highly specific selection for single-chain antibodies directed against functionally regulated epitopes on the cell surface molecules that can be tailored for diagnostic and therapeutic applications. Using this protocol, activation-specific single-chain antibodies can be obtained within 4–6 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
05 June 2008
In the version of this article initially published, some reagents were referred to incorrectly or unclearly. In Step 11, “1 ml buffer” should have been “1 ml HEPES buffer”. In Step 48, “saccharose” should have been “sucrose”. In Box 2, Step 1, “LBG media” should have been “LB media”. Some units were listed incorrectly. In Box 1, Step 3, “1,000 g ml-1” should have been “100 μg ml-1”. In the table in Step 42, the four instances of “ml” should have been “μl”. In Box 2, Step 2, “Inoculate 5 ml LB media with 100 μl” should have been “Inoculate 500 ml LB media with 5 ml”. The relationship between Procedure steps and Figure 4 was unclear, leaving the figure open to misinterpretation. The following sentence has been added to the end of Step 53, before the Critical Step: “Figure 4 shows the successful production and purification of an scFv antibody as assessed by SDS–polyacrylamide gel electrophoresis and western blotting of an scFv cloned into the pHOG-21 vector expressed in TG-1 E. coli (Steps 58–70).” The two labels on Figure 4 that read “36,000 Da” have been changed to “scFv”. In the description of panel b in the legend, “anti-HIS-tag” has been changed to “anti-His-tag”. In Step 15, “XL-1 blue on 5 ml” should have been “XL-1 blue in 5 ml”. In the legend to Figure 3, the citation to ref. 18 should have been to ref. 5. At the end of the legend to Figure 4 and in “Anticipated Results,” the citation “18,20” should have been “4–6”. These errors have been corrected in the HTML and PDF versions of the article.
References
Clackson, T., Hoogenboom, H.R., Griffiths, A.D. & Winter, G. Making antibody fragments using phage display libraries. Nature 352, 624–628 (1991).
Giordano, R.J., Cardó-Vila, M., Lahdenranta, J., Pasqualini, R. & Arap, W. Biopanning and rapid analysis of selective interactive ligands. Nat. Med. 7, 1249–1253 (2001).
Arap, W. et al. Steps toward mapping the human vasculature by phage display. Nat. Med. 8, 121–127 (2002).
Schwarz, M. et al. Single-chain antibodies for the conformation-specific blockade of activated platelet integrin alphaIIbbeta3 designed by subtractive selection from naive human phage libraries. FASEB J. 1704–1706 (2004).
Eisenhardt, S.U. et al. Generation of activation-specific human anti-alphaM/beta2 single-chain antibodies as potential diagnostic tools and therapeutic agents. Blood 109, 3521–3528 (2007).
Schwarz, M. et al. Conformation-specific blockade of the integrin GPIIb/IIIa: a novel antiplatelet strategy that selectively targets activated platelets. Circ. Res. 99, 25–33 (2006).
Fuchs, P., Breitling, F., Dübel, S., Seehaus, T. & Little, M. Targeting recombinant antibodies to the surface of Escherichia coli: fusion to a peptidoglycan associated lipoprotein. Biotechnology 9, 1369–1372 (1991).
Zahnd, C., Amstutz, P. & Plückthun, A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4, 269–279 (2007).
Roberts, R.W. & Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 94, 12297–12302 (1997).
Boder, E.T. & Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Barbas, C.F. III, Kang, A.S., Lerner, R.A. & Benkovic, S.J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88, 7978–7982 (1991).
Hust, M., Maiss, E., Jacobsen, H.J. & Reinard, T. The production of a genus-specific recombinant antibody (scFv) using a recombinant potyvirus protease. J. Virol. Methods 106, 225–233 (2002).
Hawlisch, H. et al. Site-specific anti-C3a receptor single-chain antibodies selected by differential panning on cellulose sheets. Anal. Biochem. 293, 142–145 (2001).
Moghaddam, A. et al. Identification of scFv antibody fragments that specifically recognise the heroin metabolite 6-monoacetylmorphine but not morphine. J. Immunol. Methods 280, 139–155 (2003).
Robert, R. et al. Identification of human scFvs targeting atherosclerotic lesions: selection by single round in vivo phage display. J. Biol. Chem. 281, 40135–40143 (2006).
Little, M. et al. Generation of a large complex antibody library from multiple donors. J. Immunol. Methods 231, 3–9 (1999).
Dörsam, H. et al. Antibodies to steroids from a small human naïve IgM library. FEBS Lett. 414, 7–13 (1997).
Takagi, J. & Springer, T.A. Integrin activation and structural rearrangement. Immunol. Rev. 186, 141–163 (2002).
Hynes, R.O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69, 11–25 (1992).
Eisenhardt, S.U. et al. Generation of activation-specific human anti-alphaMbeta2 single-chain antibodies as potential diagnostic tools and therapeutic agents. Blood 109, 3521–3528 (2007).
Parsons, H.L. et al. Directing phage selections towards specific epitopes. Protein Eng. 9, 1043–1049 (1996).
Osbourn, J.K., Derbyshire, E.J., Vaughan, T.J., Field, A.W. & Johnson, K.S. Pathfinder selection: in situ isolation of novel antibodies. Immunotechnology 3, 293–302 (1998).
Osbourn, J.K. et al. Directed selection of MIP-1 alpha neutralizing CCR5 antibodies from a phage display human antibody library. Nat. Biotechnol. 16, 778–781 (1998).
Levitan, B. Stochastic modeling and optimization of phage display. J. Mol. Biol. 277, 893–916 (1998).
de Wildt, R.M., Mundy, C.R., Gorick, B.D. & Tomlinson, I.M. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat. Biotechnol. 18, 989–994 (2000).
Le Gall, F., Kipriyanov, S.M., Moldenhauer, G. & Little, M. Di-, tri- and tetrameric single chain Fv antibody fragments against human CD19: effect of valency on cell binding. FEBS Lett. 453, 164–168 (1999).
Kipriyanov, S.M., Moldenhauer, G. & Little, M. High level production of soluble single chain antibodies in small-scale Escherichia coli cultures. J. Immunol. Methods 200, 69–77 (1997).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Eisenhardt, S., Schwarz, M., Bassler, N. et al. Subtractive single-chain antibody (scFv) phage-display: tailoring phage-display for high specificity against function-specific conformations of cell membrane molecules. Nat Protoc 2, 3063–3073 (2007). https://doi.org/10.1038/nprot.2007.455
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.455
This article is cited by
-
Identification of a novel fully human anti-toxic shock syndrome toxin (TSST)-1 single-chain variable fragment antibody averting TSST-1-induced mitogenesis and cytokine secretion
BMC Biotechnology (2022)
-
P2Y12 receptor blockers are anti-inflammatory drugs inhibiting both circulating monocytes and macrophages including THP-1 cells
Scientific Reports (2021)
-
A DARPin targeting activated Mac-1 is a novel diagnostic tool and potential anti-inflammatory agent in myocarditis, sepsis and myocardial infarction
Basic Research in Cardiology (2021)
-
Optimization of Tris/EDTA/Sucrose (TES) periplasmic extraction for the recovery of functional scFv antibodies
AMB Express (2020)
-
Targeting c-Met on gastric cancer cells through a fully human fab antibody isolated from a large naive phage antibody library
DARU Journal of Pharmaceutical Sciences (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.