Abstract
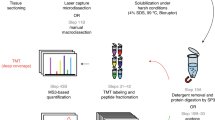
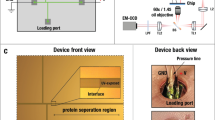
Proteome data combined with histopathological information provides important, novel clues for understanding cancer biology and reveals candidates for tumor markers and therapeutic targets. We have established an application of a highly sensitive fluorescent dye (CyDye DIGE Fluor saturation dye), developed for two-dimensional difference gel electrophoresis (2D-DIGE), to the labeling of proteins extracted from laser microdissected tissues. The use of the dye dramatically decreases the protein amount and, in turn, the number of cells required for 2D-DIGE; the cells obtained from a 1 mm2 area of an 8–12 μm thick tissue section generate up to 5,000 protein spots in a large-format 2D gel. This protocol allows the execution of large-scale proteomics in a more efficient, accurate and reproducible way. The protocol can be used to examine a single sample in 5 d or to examine hundreds of samples in large-scale proteomics.
NOTE: In the PDF version of this article initially published online, the publication date was shown as 25 January 2006 instead of 25 January 2007. The error has been corrected in the PDF version of the article.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
22 February 2007
changed 2006 to 2007
References
Espina, V. et al. Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006).
Buckanovich, R.J. et al. Use of immuno-LCM to identify the in situ expression profile of cellular constituents of the tumor microenvironment. Cancer Biol. Ther. 5, 635–642 (2006).
Banks, R.E. et al. The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis—preliminary findings. Electrophoresis 20, 689–700 (1999).
Gorg, A., Weiss, W. & Dunn, M.J. Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 (2004).
Carrette, O., Burkhard, P.R., Sanchez, J.C. & Hochstrasser, D.F. State-of-the-art two-dimensional gel electrophoresis: a key tool of proteomics research. Nat. Protoc. 1, 812–823 (2006).
Oh, J.M. et al. A database of protein expression in lung cancer. Proteomics 1, 1303–1319 (2001).
Craven, R.A., Totty, N., Harnden, P., Selby, P.J. & Banks, R.E. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am. J. Pathol. 160, 815–822 (2002).
Bravo, R., Bellatin, J. & Celis, J.E. [35S]-methionine labelled polypeptides from HELA cells. Coordinates and percentage of some major polypeptides. Cell Biol. Int. Rep. 5, 93–96 (1981).
Poznanovic, S. et al. Differential radioactive proteomic analysis of microdissected renal cell carcinoma tissue by 54 cm isoelectric focusing in serial immobilized pH gradient gels. J. Proteome Res. 4, 2117–2125 (2005).
Kondo, T. et al. Application of sensitive fluorescent dyes in linkage of laser microdissection and two-dimensional gel electrophoresis as a cancer proteomic study tool. Proteomics 3, 1758–1766 (2003).
Unlu, M., Morgan, M.E. & Minden, J.S. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 (1997).
Shaw, J. et al. Evaluation of saturation labelling two-dimensional difference gel electrophoresis fluorescent dyes. Proteomics 3, 1181–1195 (2003).
Lee, J.R. et al. Differential protein analysis of spasomolytic polypeptide expressing metaplasia using laser capture microdissection and two-dimensional difference gel electrophoresis. Appl. Immunohistochem. Mol. Morphol. 11, 188–193 (2003).
Zhou, G. et al. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol. Cell Proteomics 1, 117–124 (2002).
Alban, A. et al. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3, 36–44 (2003).
Uwakwe, C.J. CyDye DIGE Fluor saturation dyes for labelling and detection of scarce protein samples using Ettan DIGE system: application of the pooled internal standard to the investigation of a model system. Amersham Biosciences Life Science News 16, 1–4 (2004).
Uhlen, M. & Ponten, F. Antibody-based proteomics for human tissue profiling. Mol. Cell Proteomics 4, 384–393 (2005).
Okano, T. et al. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE). Proteomics 6, 3938–3948 (2006).
Tan, H.T., Zubaidah, R.M., Tan, S., Hooi, S.C. & Chung, M.C. 2-D DIGE analysis of butyrate-treated HCT-116 cells after enrichment with heparin affinity chromatography. J. Proteome Res. 5, 1098–1106 (2006).
Ahmed, N. & Rice, G.E. Strategies for revealing lower abundance proteins in two-dimensional protein maps. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 815, 39–50 (2005).
Lescuyer, P., Hochstrasser, D.F. & Sanchez, J.C. Comprehensive proteome analysis by chromatographic protein prefractionation. Electrophoresis 25, 1125–1135 (2004).
Qin, S. et al. Chromatofocusing fractionation and two-dimensional difference gel electrophoresis for low abundance serum proteins. Proteomics 5, 3183–3192 (2005).
Young, D.A., Voris, B.P., Maytin, E.V. & Colbert, R.A. Very-high-resolution two-dimensional electrophoretic separation of proteins on giant gels. Meth. Enzymol. 91, 190–214 (1983).
Klose, J. et al. Genetic analysis of the mouse brain proteome. Nat. Genet. 30, 385–393 (2002).
Seike, M. et al. Proteomic signatures for histological types of lung cancer. Proteomics 5, 2939–2948 (2005).
Fujii, K. et al. Proteomic study of human hepatocellular carcinoma using two-dimensional difference gel electrophoresis with saturation cysteine dye. Proteomics 5, 1411–1422 (2005).
Fujii, K. et al. Database of two-dimensional polyacrylamide gel electrophoresis of proteins labeled with CyDye DIGE Fluor saturation dye. Proteomics 6, 1640–1653 (2006).
Suehara, Y. et al. Proteomic signatures corrsponding to histological classification and grading of soft-tissue sarcomas. Proteomics 6, 4402–4409 (2006).
Hatakeyama, H. et al. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics 6, 6300–6317 (2005).
Rhodes, D.R. & Chinnaiyan, A.M. Integrative analysis of the cancer trasncriptome. Nat. Genet. 37, S31–S37 (2005).
Chen, G. et al. Protein profiles associated with survival in lung adenocarcinoma. Proc. Natl Acad. Sci. USA 100, 13537–13542 (2003).
Greengauz-Roberts, O. et al. Saturation labeling with cysteine-reactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics 5, 1746–1757 (2005).
Wilson, K.E. et al. Comparative proteomic analysis using samples obtained with laser microdissection and saturation dye labelling. Proteomics 5, 3851–3858 (2005).
Sitek, B. et al. Application of fluorescence difference gel electrophoresis saturation labelling for the analysis of microdissected precursor lesions of pancreatic ductal adenocarcinoma. Proteomics 5, 2665–2679 (2005).
Sitek, B. et al. Novel approaches to analyse glomerular proteins from smallest scale murine and human samples using DIGE saturation labelling. Proteomics 6, 4337–4345 (2006).
GE Healthcare Bio-Sciences. Detection and mass spectrometry identification of protein changes in low-abundance tissue using CyDye DIGE Fluor saturation dyes. Application note 18117751AA, 2003.10, 1–8.
Rabilloud, T., Valette, C. & Lawrence, J.J. Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis 15, 1552–1558 (1994).
Sanchez, J.C. et al. Improved and simplified in-gel sample application using reswelling of dry immobilized pH gradients. Electrophoresis 18, 324–327 (1997).
Sapan, C.V., Lundblad, R.L. & Price, N.C. Colorimetric protein assay techniques. Biotechnol. Appl. Biochem. 29, 99–108 (1999).
Maeda, K., Finnie, C. & Svensson, B. Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem. J. 378, 497–507 (2004).
Fujii, K. et al. Protein expression pattern distinguishes different lymphoid neoplasms. Proteomics 5, 4274–4286 (2005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Kondo, T., Hirohashi, S. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat Protoc 1, 2940–2956 (2006). https://doi.org/10.1038/nprot.2006.421
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.421
This article is cited by
-
CDK1 dependent phosphorylation of hTERT contributes to cancer progression
Nature Communications (2020)
-
Establishment and characterization of a novel cell line, NCC-MFS1-C1, derived from a patient with myxofibrosarcoma
Human Cell (2019)
-
Establishment of novel patient-derived models of dermatofibrosarcoma protuberans: two cell lines, NCC-DFSP1-C1 and NCC-DFSP2-C1
In Vitro Cellular & Developmental Biology - Animal (2019)
-
Differential regulation of cysteine oxidative post-translational modifications in high and low aerobic capacity
Scientific Reports (2018)
-
Establishment and proteomic characterization of a novel synovial sarcoma cell line, NCC-SS2-C1
In Vitro Cellular & Developmental Biology - Animal (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.