Abstract
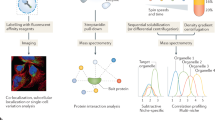
As proteins within cells are spatially organized according to their role, knowledge about protein localization gives insight into protein function. Here, we describe the LOPIT technique (localization of organelle proteins by isotope tagging) developed for the simultaneous and confident determination of the steady-state distribution of hundreds of integral membrane proteins within organelles. The technique uses a partial membrane fractionation strategy in conjunction with quantitative proteomics. Localization of proteins is achieved by measuring their distribution pattern across the density gradient using amine-reactive isotope tagging and comparing these patterns with those of known organelle residents. LOPIT relies on the assumption that proteins belonging to the same organelle will co-fractionate. Multivariate statistical tools are then used to group proteins according to the similarities in their distributions, and hence localization without complete centrifugal separation is achieved. The protocol requires approximately 3 weeks to complete and can be applied in a high-throughput manner to material from many varied sources.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Millar, A.H. Location, location, location: surveying the intracellular real estate through proteomics in plants. Funct. Plant Biol. 31, 20 (2004).
Dupree, P. & Sherrier, D.J. The plant Golgi apparatus. Biochem. Biophys. Acta Mol. Cell Res. 1404, 259–270 (1998).
Galili, G., Sengupta-Gopalan, C. & Ceriotti, A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol. Biol. 38, 1–29 (1998).
Eisenhaber, F. & Bork, P. Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol. 8, 169–170 (1998).
Nakai, K. & Horton, P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35 (1999).
Corse, E. & Machamer, C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76, 1273–1284 (2002).
Davis, T.N. Protein localization in proteomics. Curr. Opin. Chem. Biol. 8, 49–53 (2004).
Lippincott-Schwartz, J. & Patterson, G.H. Development and use of fluorescent protein markers in living cells. Science 300, 87–91 (2003).
Mura, C.V., Becker, M.I., Orellana, A. & Wolff, D. Immunopurification of Golgi vesicles by magnetic sorting. Immunol. Methods 260, 263–71 (2002).
Taylor, S.W. et al. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 (2003).
Friso, G. et al. In depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499 (2004).
Wu, C.C. et al. Organellar proteomics reveals Golgi arginine dimethylation. Mol. Biol. Cell 15, 2907–2919 (2004).
Jurgens, G. Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20, 481–504 (2004).
Dunkley, T.P., Watson, R., Griffin, J.L., Dupree, P. & Lilley, K.S. Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell Proteomics 3, 1128–1134 (2004).
Dunkley, T.P. et al. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103, 6518–6523 (2006).
Ross, P.L. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics 3, 1154–1169 (2004).
Andersen, J.S. et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 (2003).
Foster, L. et al. A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 (2006).
Tan, D.J. & Martinez-Arias, A. High-throughput localization of organelle proteins by mass spectrometry: a quantum leap for cell biology. BioEssays 28, 780–784 (2006).
Prime, T.A., Sherrier, D.J., Mahon, P., Packman, L.C. & Dupree, P. A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21, 3488–3499 (2000).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-encoded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Berg, M., Parbel, A., Pettersen, H., Feny, D. & Björkesten, L. Reproducibility of LC–MS-based protein identification. J. Exp. Bot. 57, 1509–1514 (2006).
Boehm, A.M., Galvin, R.P. & Sickmann, A. Extractor for ESI quadrupole TOF tandem MS data enabled for high throughput batch processing. BMC Bioinformatics 5, 162 (2004).
Shadforth, I.P., Xu, W., Crowther, D. & Bessant, C. GAPP: A fully automated software for the confident identification of human peptides from tandem mass spectra. J. Proteome Res. 5, 2849–2852 (2006).
Shadforth, I.P., Dunkley, T.P., Lilley, K.S. & Bessant, C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6, 145 (2005).
Graham, J., Ford, T. & Rickwood, D. The preparation of subcellular organelles from mouse-liver in self-generated gradients of iodixanol. Anal. Biochem. 220, 367–373 (1994).
Fujiki, Y., Hubbard, A.L., Fowler, S. & Lazarow, P.B. Isolation of intracellular membranes by means of sodium-carbonate treatment. J. Cell Biol. 93, 97–102 (1982).
Shadforth, I.P., Dunkley, T.P., Lilley, K., Crowther, D. & Bessant, C. Confident protein identification using the average peptide score method coupled with search-specific, ab initio, thresholds. Rapid Commun. Mass Spectrom. 19, 3363–3368 (2005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Sadowski, P., Dunkley, T., Shadforth, I. et al. Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 1, 1778–1789 (2006). https://doi.org/10.1038/nprot.2006.254
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.254
This article is cited by
-
Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics
Nature Communications (2019)
-
Using hyperLOPIT to perform high-resolution mapping of the spatial proteome
Nature Protocols (2017)
-
Enhancing gold recovery from electronic waste via lixiviant metabolic engineering in Chromobacterium violaceum
Scientific Reports (2013)
-
Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods
The EMBO Journal (2011)
-
Generating and navigating proteome maps using mass spectrometry
Nature Reviews Molecular Cell Biology (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.