Abstract
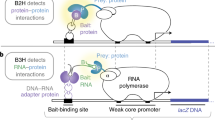
We describe an in-cell NMR-based method for mapping the structural interactions (STINT-NMR) that underlie protein-protein complex formation. This method entails sequentially expressing two (or more) proteins within a single bacterial cell in a time-controlled manner and monitoring their interactions using in-cell NMR spectroscopy. The resulting NMR data provide a complete titration of the interaction and define structural details of the interacting surfaces at atomic resolution. Unlike the case where interacting proteins are simultaneously overexpressed in the labeled medium, in STINT-NMR the spectral complexity is minimized because only the target protein is labeled with NMR-active nuclei, which leaves the interactor protein(s) cryptic. This method can be combined with genetic and molecular screens to provide a structural foundation for proteomic studies. The protocol takes 4 d from the initial transformation of the bacterial cells to the acquisition of the NMR spectra.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gerstein, M., Lan, N. & Jansen, R. Proteomics. Integrating interactomes. Science 295, 284–287 (2002).
Jansen, R., Lan, N., Qian, J. & Gerstein, M. Integration of genomic datasets to predict protein complexes in yeast. J. Struct. Funct. Genomics 2, 71–81 (2002).
Uetz, P. & Hughes, R.E. Systematic and large-scale two-hybrid screens. Curr. Opin. Microbiol. 3, 303–308 (2000).
Burz, D.S., Dutta, K., Cowburn, D. & Shekhtman, A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 3, 91–93 (2006).
Shekhtman, A., Ghose, R., Goger, M. & Cowburn, D. NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett. 524, 177–182 (2002).
Serber, Z. & Dotsch, V. In-cell NMR spectroscopy. Biochemistry 40, 14317–14323 (2001).
Lutz, R., Lozinski, T., Ellinger, T. & Bujard, H. Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator. Nucleic Acids Res. 29, 3873–3881 (2001).
Zuger, S. & Iwai, H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat. Biotechnol. 23, 736–740 (2005).
Groll, M., Bochtler, M., Brandstetter, H., Clausen, T. & Huber, R. Molecular machines for protein degradation. Chembiochem 6, 222–256 (2005).
Riek, R., Pervushin, K. & Wuthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 25, 462–468 (2000).
Piotto, M., Saudek, V. & Sklenar, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol. NMR 2, 661–665 (1992).
Zuiderweg, E.R. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 41, 1–7 (2002).
Wuthrich, K. NMR of Proteins and Nucleic Acids 8–10 (John Wiley & Sons, New York, 1986).
Guex, N. & Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Acknowledgements
This work was supported by grants to A.S. (American Diabetes Association Career Development Award) and to D.C. (National Institutes of Health).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Burz, D., Dutta, K., Cowburn, D. et al. In-cell NMR for protein-protein interactions (STINT-NMR). Nat Protoc 1, 146–152 (2006). https://doi.org/10.1038/nprot.2006.23
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.23
This article is cited by
-
Caught in Action: Selecting Peptide Aptamers Against Intrinsically Disordered Proteins in Live Cells
Scientific Reports (2015)
-
NMR protein structure determination in living E. coli cells using nonlinear sampling
Nature Protocols (2010)
-
Structural determination of biomolecular interfaces by nuclear magnetic resonance of proteins with reduced proton density
Journal of Biomolecular NMR (2010)
-
Site-specific labeling of proteins with NMR-active unnatural amino acids
Journal of Biomolecular NMR (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.