Abstract
The bacterium Caulobacter crescentus swims by rotating a single right-handed helical filament. These cells have two swimming modes: a pusher mode, in which clockwise (CW) rotation of the filament thrusts the cell body forwards1, and a puller mode, in which counterclockwise (CCW) rotation pulls it backwards2. The situation is reversed in Escherichia coli, a bacterium that rotates several left-handed filaments CCW to drive the cell body forwards. The flagellar motor in E. coli generates more torque in the CCW direction than the CW direction in swimming cells3,4. However, C. crescentus and other bacteria with single filaments swim forwards and backwards at similar speeds, prompting the assumption that motor torques in the two modes are the same5,6. Here, we present evidence that motors in C. crescentus develop higher torques in the puller mode than in the pusher mode, and suggest that the anisotropy in torque generation is similar in the two species, despite the differences in filament handedness and motor bias.
Similar content being viewed by others
Main
The differences in the fluid flows generated in the pusher and puller modes can be approximated by force dipoles of opposite signs but equal strengths, and the assumption of equal motor torques is important when comparing the hydrodynamics and rheology of active fluids7,8,9,10,11. Given this assumption, that the cells in Vibrio alginolyticus swim twice as fast backwards as forwards, only when close to solid surfaces, has been attributed to changes in swimming geometries and hydrodynamic interactions with the bounding surfaces12,13. However, such mechanisms fail to explain why cells in Pseudomonas putida swim twice as fast backwards at large separations from surfaces14.
We observed cell bodies that transiently tethered to a glass surface and rotated about an axis normal to the surface. Such cells rotated twice as fast CW as CCW. The filaments remained untethered and rotated freely, as shown by visualization of fluorescently labelled flagella. The cell-body rotation could be understood on the basis of a model that took into account the hydrodynamic interactions between a rolling filament and the underlying surface. The model predicted that CW rotation of a filament drives the tethered cell CCW, and CCW rotation of the filament drives the tethered cell CW. Thus, under moderate viscous loads, the filaments in C. crescentus rotate faster in the CCW direction than in the CW direction. We tested the model by calculating the fraction of the time that motors rotate CW (motor bias or CWbias) and determined that the CWbias was 0.8. This was consistent with the observations that C. crescentus swam forwards most of the time. The anisotropy in motor speeds disappeared at very high viscous loads, similar to the motor behaviour in E. coli3,4. On the basis of these results, we assert that the anisotropy in torque generation is similar in the two species, although filament handedness and motor bias are opposite.
In our experiments, swimming cells of C. crescentus strains lacking pili spontaneously stuck to the microscope coverglass, and rotated in either direction about a single point of attachment. Most cells would detach and swim away after ∼1–5 min. Digital video recordings were analysed as before15 to track the lengths/widths of the cells as they appeared in the image plane, their orientations, the centre of the cell body and the point of tether (centre of rotation) over time (Methods). Two types of cell rotations about the z-axis (the axis normal to the image plane) were observed. In the first case, the cell would remain in-plane when rotating CCW, but would orient vertically when rotating CW, as shown schematically in Fig. 1a. In this case, the centre of the cell-body exhibited concentric circular trajectories (bottom panel)—the inner trajectory for CW rotation and the outer trajectory for CCW rotation, as shown in Fig. 1a. As the hydrodynamic drag on the body was different in these two geometries, we did not include such trajectories for further analysis of motor torques. In the second case, the cell rotated in-plane (Fig. 1b) and the centre of the cell body exhibited a single circular trajectory irrespective of the direction of rotation (bottom panel). Only such cells were selected for further analysis.
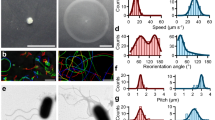
a, First type of tethering observed in C. crescentus. Rotation geometry (top panel). CCW cell rotation is indicated by the grey cell and CW cell rotation is indicated by the white cell (dashed outline). The black dots show the respective centres of body tracked by the algorithm. The corresponding trajectories traced by the respective black dots are indicated (bottom panel). b, Second type of tethering observed in C. crescentus. Rotation geometry (top panel). The black dot indicates the centre of body tracked by the tracking algorithm and the white circle indicates the centre of rotation. The corresponding trajectories traced by the centre of body are indicated (bottom panel).
A typical trace of rotation speed, Ω, calculated from the change in cell orientation with time, is shown in Fig. 2a. The magnitudes of the speeds showed a clear difference in the two directions. To quantitatively determine if the variations in speed of rotation were due to changes in tethering geometry, we analysed the lengths (L) and widths (W) of the cells (projected on the image plane) over time. For the cell shown in Fig. 2a, L versus time is indicated in Fig. 2b over the same time interval. The length of the tracked cell seemed to remain constant over the duration of our measurement. No significant cross-correlation was observed between either L and Ω (inset in Fig. 2b) or W and Ω (not shown). The point of tether or the centre of rotation (see schematic in Fig. 1b and Supplementary Fig. 1) also remained unchanged. This indicated that the tethering geometry remained the same in either direction. The distribution of the ratios of the absolute cell-rotation speeds, ΩCW versus ΩCCW, is plotted in Fig. 2c (n = 48 motors). The average ratio (ΩCW/ΩCCW) was 2.2 ± 0.9 and the average CW cell-rotation speed was ∼−8 Hz.
a, Typical cell-body rotational speeds over time. Note that the CCW cell-rotation speed (positive values) is lower than the absolute CW cell-rotation speed (negative values). b, Trace of the length, L, of the cell shown in a projected on the image plane during the same interval. Inset shows the cross-correlation between Ω and L. c, Distributions of the absolute value of the ratio of cell-rotation speeds (n = 48 cells).
The cell body is crescent-shaped. As a result, any significant rotation of the cell about its principal axis (which lies parallel to the surface) would be detectable through changes in the shape of the cell image. Given the asymmetry in the cell shape, changes in the orientation along the principal axis during a switch in the direction of cell rotation could affect the relative drag coefficients in the two directions. However, no obvious changes in the shape of the cell image were observed regardless of the direction, in the manner expected for a cell rotating about an axis normal to its principal axis (Supplementary Movie 1, 0.75 × speed). Furthermore, experiments with straight mutants, which are symmetric along the principal axis16, revealed a anisotropy in relative speeds similar to that seen in Fig. 2c. Thus, variations in the relative drag coefficients of the cell body make insignificant contributions to the anisotropy in rotational speeds.
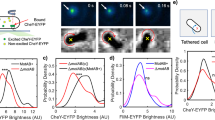
To determine the mechanism of rotation, we fluorescently labelled the flagellar filaments and visualized rotating cells under total internal reflection fluorescence (TIRF) illumination (see Methods). Figure 3a and Supplementary Movie 2 show the rotation of a cell with a labelled flagellum. As is evident, the filament goes around with the cell body, rotating about an axis extending from one end of the cell. The cells were not tethered by their flagellar filaments. The tethering point lies on the surface of the cell body, an attachment probably due to nonspecific interactions between the cell surface and the surface. We refer to the attachment as a fluid joint, which is probably made up of the polysaccharide that covers the entire cell in C. Crescentus17. Further discussions on the tether properties have been included in the Supplementary Information (see Supplementary Figs 2 and 3).
a, Visualization of fluorescently labelled flagellar filaments. The bright spot indicates the flagellum and the dashed ellipses the cell body. Direction of rotation of the cell body is indicated by the white arrow. b, Model for cell rotation: CCW rotation of the flagellum causes CW cell rotation through hydrodynamic coupling with the solid substrate. c, Thrust-based mechanisms are unlikely to explain the results in Fig. 2c as, depending on the relative position of the tethering point, the same force F can cause CCW cell rotation (in case of the white tethering point) or CW rotation (in case of the grey tethering point). d, The fraction of time that motors spin CW (the CWbias calculated on the basis of the model shown in b) shows a peak in distribution around 0.8, consistent with previous measurements that show that cells swim forwards a majority of the time.
As seen in the movie, the filament is moving sideways. We believe this lateral force is generated by hydrodynamic coupling of the flagellar filament with the underlying surface, as the helix rolls over that surface. To understand this effect, consider a rotating sphere in place of the filament. As the drag on its bottom surface is higher than the drag on the top surface, the sphere will translate laterally owing to its rotation. This mechanism is responsible for the spiralling of E. coli as it swims near a substrate18. When C. crescentus rotates over the surface (Fig. 3b) the filament rolls, but the cell body is not able to do so; it is constrained by the fluid joint that tethers it to the surface. It is important to note that when the filament spins CW (when viewed from its distal end), the body turns CCW (when viewed from above). Because the cells rotate about twice as fast CW as CCW (Fig. 2), the filaments must rotate about twice as fast CCW as CW: the CCW motor torque is larger than the CW torque.
Other mechanisms could involve the thrust (F) generated by the rotation of the filament. For a finite distance r between the helix axis and the tethering point, a torque τ(∼r × F) could cause the cell to rotate as shown in Fig. 3c. However, r is typically small (∼100 nm) and hence the thrust would be inadequate to rotate a cell at 4–8 Hz. Furthermore, such a mechanism can cause cell rotation in either direction at different speeds, depending on the point of tethering (Fig. 3c) and the orientation of the filament (with respect to the cell’s principal axis). However, faster CW rotation was predominantly observed in our data. Evidently, such mechanisms do not contribute much to cell rotation (see Supplementary Text).
A fraction of cells (<1%) were observed to tether and rotate in such a way that they remained perpendicular to the surface in either direction, with the filaments pointing away from the surface. Flagellar interactions with the surface were absent and the cells counter-rotated owing to motor rotation. A similar anisotropy in the speeds of cell rotation was observed (ΩCW ∼ 1.5 ΩCCW), consistent with the hypothesis that motors rotate faster in the CCW direction (Supplementary Fig. 3). Next, we irreversibly tethered the filament to the surface, which resulted in high loads on the motor. Consistent with previous reports in E. coli, the anisotropy in speeds vanished at these higher loads (Supplementary Fig. 4).
To test our model, we determined the CWbias. The inversion between cell and filament rotational directions is a direct consequence of the hydrodynamic interactions with the surface, as discussed above. Therefore, the CWbias is simply the fraction of time that the cell body rotates CCW. The observed CWbias is shown in Fig. 3d and has an average value of 0.8. Our model thus predicts that the cells swim forwards 80% of the time, which is consistent with our observations of swimming cells with fluorescently labelled flagella and those of Liu and co-workers6. This supports the hypothesis of rotational direction-inversion.
Recent measurements by Liu et al. 6 indicated that the body counter-rotation frequency is twice as high in the puller mode in C. crescentus cells that swim in the bulk fluid. This difference was attributed to differences in the kinematics of swimming in the two directions, because the cells precess along a helical path of higher amplitude in the pusher mode than in the puller mode. Paradoxically, the swimming speeds in the two modes are identical, which indicates that the thrust developed is independent of the direction. On the basis of our results, we suggest that the torque generated by the motor in the CCW direction and, consequently, the filament rotation frequency, is higher when the cell swims backwards (ωpuller ∼ 1.5–2 times ωpusher). Therefore, the counter-rotation frequency in the puller mode is larger as well (Ωpuller ∼ 1.5–2 times Ωpusher). We extended the precession model6 to explain why the overall thrust developed, and hence the swimming speed (V), is independent of the direction of motor rotation. At low Reynolds numbers, the following relations apply for the right-handed helix:

whereas, for the cell, the following relations hold:

The non-diagonal elements of the resistance tensor Yij are non-zero because the cells precess (along a left-handed helical path). The slantwise motion of the cell body during precession develops thrust, adding to that developed by the filament, compensating for the lower motor torque in the pusher mode. The right-hand rule was adopted in determining the sign of ω, and the signs on the elements of Yij were adjusted taking into account the opposite directions of body counter-rotation and filament rotation. For force-free and torque-free swimming cells, Fcell + Ffil = 0 and τcell + τfil = 0. Therefore equations ((1)) and ((2)) can be solved to obtain the expression for the ratio of swimming speeds (VB/VA) in cases A and B, for example when ωB = 2ωA and ΩB = 2ΩA.

The values of the elements of Yij change with the amplitude of precession6. Xij was calculated using Lighthill’s slender-body theory19,20. First, we assumed that the elements of the resistance tensor Xij are invariant with respect to the direction of rotation, as we did not observe significant changes in the polymorphic form of the filament (Supplementary Fig. 5). During precession, the cell’s principal axis makes an angle with the direction of swimming, defined as the angle of precession (θ; ref. 6). Assuming an angle of precession θB ∼ 0.25 rad, we calculated the ratios from equation (3) for varying θA, as shown in Fig. 4a. For θA ∼ 0.5 rad, swimming speeds for the two cases are equal for all cell lengths analysed (1.25, 2 and 2.75 μm), even though the filament rotation frequency is twice as large in case B.
a, Predicted ratios of swimming speeds (VB/VA) when the filament and cell-body rotational frequencies differ in the two cases (ωB = 2ωA and ΩB = 2ΩA). Precession angle θB = 0.25 rad. b, Effect of change in filament shape with direction of rotation. Precession angle θB = 0.35 rad, cell length L = 2.5 μm and RCW = 0.13 μm. All predictions are from equation (3).
Next, we considered the effect of a change in the shape of the filament with direction of rotation, such that XijCCW ≠ XijCW. Assuming a constant arc length, Λ, we varied the radius of the helix for CCW turns (RCCW) and determined the pitch (λCCW) from the relation Λ = nc(4π2RCCW2 + λCCW2)0.5, where nc is the number of helical turns. The dependence of VB/VA on the values of precession angles and the shape of the filament are shown in Fig. 4b (for RCW = 0.13 μm and θB = 0.35 rad). As evident, even a ∼20% change in filament helical radius does not significantly alter the ratios. Thus, the lateral force generated by cell precession is indeed sufficient to overcome the lower motor torque in case A. Note that the values of precession angles in our analysis (θB ∼ 0.35 rad and θA ∼ 0.5 rad) are in close agreement with the values experimentally measured (θpuller ∼ 0.35 rad and θpusher ∼ 0.49 rad) by Liu et al.
The similarities in motor behaviour in C. crescentus and E. coli reported here seem consistent with the structural similarities in the flagellar motors in the two species21, despite the differences in filament handedness. In C. crescentus, flexibility in the polymorphic form of the filament will result in unidirectional motion, as a CW rotating right-handed helix and a CCW rotating left-handed helix generate thrust in the same direction. One way to prevent such changes in form is to increase the structural integrity by adding multiple flagellins as reinforcements. This could be the underlying reason for the redundancy in flagellin genes in C. crescentus22. Finally, C. crescentus has an asymmetric developmental cycle23, in which a cell irreversibly tethers to a substrate and releases a new swimmer cell after cell division. It is likely that the higher torque in the puller mode helps the swimmer cell detach quickly from its mother.
Methods
Strains and plasmids.
We used a ΔpilA strain (NA1000) lacking the holdfast for all experiments. For TIRF visualization, site-directed mutagenesis was conducted on the fljK gene to change the codon encoding alanine125 to a codon encoding cysteine125. The construct was then cloned in pBXMCS-2 (ref. 24) using the restriction sites NdeI and XbaI. The plasmid was transformed into strain TPA12-ΔfljJ-K-L-M-N-O (ref. 22) and expression was induced by adding Xylose (∼0.08 M) at the start of the growth culture. All strains were grown in peptone-yeast extract (PYE) at 30 °C, and kanamycin (20 μg ml−1) was added wherever necessary at the start of the growth culture. To label the filaments fluorescently, we used an Alexa Fluor dye (532 nm, maleimide derivative, Life Technologies).
Rotational speed measurements and TIRF imaging.
Cells were introduced in tunnel slides and cell rotation was recorded using a digital camera (Thorlabs DCC1240M) at 67 frames per second with a ×40 phase objective and a ×1.6 optovar setting, when cells happened to approach and tether transiently to the coverglass. Direct imaging was performed to confirm bacterial crescent-cell shape and motility. No contamination was observed on growth media. We did experiments with both kinds of coverglass, out of the box and those cleaned with alcoholic KOH. The instances of cell adhesion were higher in the former and lower in the latter case. However, in either case, CW rotation of the cell was predominantly faster. Cells that were selected for analysis rotated smoothly without pausing for a minimum of ∼1 min. To track the centre of rotation (point of tether or pivot) in either direction, we constructed single images by averaging several frames together, in which the cells rotated in only one direction. This resulted in a bright spot with a Gaussian intensity profile, the centre of which was the point of tether. We applied standard particle tracking algorithms25 to determine the centre of such Gaussian profiles. This gave us the centre of rotation for CW and CCW. To track the centre of body, which is distinct from the point of tether, we used a segmentation algorithm (MATLAB, Mathworks) and fitted ellipses to the cell profile. We then determined the cell orientation, the length/width of the cells and the centre of the cell body by fitting ellipses to such binary images. The orientation changed with respect to time, which enabled us to calculate the rotational speeds. The centre of the cell body traced out a circular trajectory, which enabled us to discriminate between the geometries discussed in Fig. 1a, b. The cross-correlation between cell lengths (L)/widths (W) and motor speeds was calculated as

Ω′ was obtained by normalizing positive Ω values with the mean absolute CCW speed, and negative Ω values were normalized with the absolute mean CW speed. L′ was obtained by centring and scaling L. The set-up for simultaneous fluorescence and phase contrast imaging for visualization of the fluorescently labelled filaments is described elsewhere26.
References
Koyasu, S. & Shirakihara, Y. Caulobacter crescentus flagellar filament has a right-handed helical form. J. Mol. Biol. 173, 125–130 (1984).
Lauga, E. & Powers, T. R. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 72, 096601 (2009).
Chen, X. B. & Berg, H. C. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys. J. 78, 1036–1041 (2000).
Yuan, J., Fahrner, K. A., Turner, L. & Berg, H. C. Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 107, 12846–12849 (2010).
Li, G. & Tang, J. X. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys. J. 91, 2726–2734 (2006).
Liu, B. et al. Helical motion of the cell body enhances Caulobacter crescentus motility. Proc. Natl Acad. Sci. USA 111, 11252–11256 (2014).
Rafai, S., Jibuti, L. & Peyla, P. Effective viscosity of microswimmer suspensions. Phys. Rev. Lett. 104, 098102 (2010).
Saintillan, D. The dilute rheology of swimming suspensions: A simple kinetic model. Exp. Mech. 50, 1275–1281 (2010).
Underhill, P. T., Hernandez-Ortiz, J. P. & Graham, M. D. Diffusion and spatial correlations in suspensions of swimming particles. Phys. Rev. Lett. 100, 248101 (2008).
Watari, N. & Larson, R. G. The hydrodynamics of a run-and-tumble bacterium propelled by polymorphic helical flagella. Biophys. J. 98, 12–17 (2010).
Hatwalne, Y., Ramaswamy, S., Rao, M. & Simha, R. A. Rheology of active-particle suspensions. Phys. Rev. Lett. 92, 118101 (2004).
Magariyama, Y. et al. Difference in bacterial motion between forward and backward swimming caused by the wall effect. Biophys. J. 88, 3648–3658 (2005).
Goto, T., Nakata, K., Baba, K., Nishimura, M. & Magariyama, Y. A fluid-dynamic interpretation of the asymmetric motion of singly flagellated bacteria swimming close to a boundary. Biophys. J. 89, 3771–3779 (2005).
Theves, M., Taktikos, J., Zaburdaev, V., Stark, H. & Beta, C. A bacterial swimmer with two alternating speeds of propagation. Biophys. J. 105, 1915–1924 (2013).
Lele, P. P., Hosu, B. G. & Berg, H. C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 110, 11839–11844 (2013).
Ausmees, N., Kuhn, J. R. & Jacobs-Wagner, C. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115, 705–713 (2003).
Seltmann, G. & Holst, O. The Bacterial Cell Wall (Springer, 2001).
Lauga, E., DiLuzio, W. R., Whitesides, G. M. & Stone, H. A. Swimming in circles: Motion of bacteria near solid boundaries. Biophys. J. 90, 400–412 (2006).
Rodenborn, B., Chen, C. H., Swinney, H. L., Liu, B. & Zhang, H. P. Propulsion of microorganisms by a helical flagellum. Proc. Natl Acad. Sci. USA 110, E338–E347 (2013).
Lighthill, J. Flagellar hydrodynamics–Neumann, JV Lecture, 1975. Soc. Ind. Appl. Math. Rev. 18, 161–230 (1976).
Stallmeyer, M. J., Hahnenberger, K. M., Sosinsky, G. E., Shapiro, L. & DeRosier, D. J. Image reconstruction of the flagellar basal body of Caulobacter crescentus. J. Mol. Biol. 205, 511–518 (1989).
Faulds-Pain, A. et al. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J. Bacteriol. 193, 2695–2707 (2011).
Shapiro, L. Differentiation in the Caulobacter cell cycle. Annu. Rev. Microbiol. 30, 377–407 (1976).
Thanbichler, M., Iniesta, A. A. & Shapiro, L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucl. Acids Res. 35, e137 (2007).
Crocker, J. C. & Grier, D. G. Methods of digital video microscopy for colloidal studies. J. Colloid Interf. Sci. 179, 298–310 (1996).
Lele, P. P., Branch, R. W., Nathan, V. S. & Berg, H. C. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc. Natl Acad. Sci. USA 109, 20018–20022 (2012).
Acknowledgements
We thank P. Aldridge, C. Jacobs-Wagner and M. Laub for strains. We are grateful to I. Hug and U. Jenal for strains, reagents and advice. The work was supported by National Institutes of Health Grant AI016478.
Author information
Authors and Affiliations
Contributions
P.P.L. and H.C.B. designed the work; P.P.L., T.R., A.S. and Y.C. performed the research; P.P.L., T.R. and Y.C. analysed the data; P.P.L., T.R. and H.C.B. developed the experimental set-up; and P.P.L. and H.C.B. wrote the paper with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1144 kb)
Supplementary Movie 1
Supplementary Movie (AVI 1778 kb)
Supplementary Movie 2
Supplementary Movie (AVI 2463 kb)
Supplementary Movie 3
Supplementary Movie (AVI 536 kb)
Supplementary Movie 4
Supplementary Movie (AVI 660 kb)
Supplementary Movie 5
Supplementary Movie (AVI 1234 kb)
Rights and permissions
About this article
Cite this article
Lele, P., Roland, T., Shrivastava, A. et al. The flagellar motor of Caulobacter crescentus generates more torque when a cell swims backwards. Nature Phys 12, 175–178 (2016). https://doi.org/10.1038/nphys3528
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nphys3528
This article is cited by
-
Linear motor driven-rotary motion of a membrane-permeabilized ghost in Mycoplasma mobile
Scientific Reports (2018)
-
Torque, but not FliL, regulates mechanosensitive flagellar motor-function
Scientific Reports (2017)
-
Experimental characterization of helical swimming trajectories in circular channels
Microfluidics and Nanofluidics (2017)