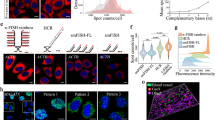
Abstract
Alternative messenger RNA (mRNA) splicing is a fundamental process of gene regulation, and errors in RNA splicing are known to be associated with a variety of different diseases. However, there is currently a lack of quantitative technologies for monitoring mRNA splice variants in cells. Here, we show that a combination of plasmonic dimer probes and hyperspectral imaging can be used to detect and quantify mRNA splice variants in living cells. The probes are made from gold nanoparticles functionalized with oligonucleotides and can hybridize to specific mRNA sequences, forming nanoparticle dimers that exhibit distinct spectral shifts due to plasmonic coupling. With this approach, we show that the spatial and temporal distribution of three selected splice variants of the breast cancer susceptibility gene, BRCA1, can be monitored at single-copy resolution by measuring the hybridization dynamics of the nanoplasmonic dimers. Our study provides insights into RNA and its transport in living cells, which could improve our understanding of cellular protein complexes, pharmacogenomics, genetic diagnosis and gene therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Orban, T. I. & Olah, E. Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem. Biophys. Res. Commun. 280, 32–38 (2001).
Budhram-Mahadeo, V., Ndisang, D., Ward, T., Weber, B. L. & Latchman, D. S. The Brn-3b POU family transcription factor represses expression of the BRCA-1 anti-oncogene in breast cancer cells. Oncogene 18, 6684–6691 (1999).
Xu, X. L. et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nature Genet. 22, 37–43 (1999).
Wilson, C. A. et al. Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-Delta 11b. Oncogene 14, 1–16 (1997).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods 5, 877–879 (2008).
Trcek, T., Larson, D. R., Moldón, A., Query, C. C. & Singer, R. H. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147, 1484–1497 (2011).
Levsky, J. M., Shenoy, S. M., Pezo, R. C. & Singer, R. H. Single-cell gene expression profiling. Science 297, 836–840 (2002).
Shav-Tal, Y. et al. Dynamics of single mRNPs in nuclei of living cells. Science 304, 1797–1800 (2004).
Santangelo, P., Nitin, N. & Bao, G. Nanostructured probes for RNA detection in living cells. Ann. Biomed. Eng. 34, 39–50 (2006).
Bratu, D. P., Cha, B-J., Mhlanga, M. M., Kramer, F. R. & Tyagi, S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA 100, 13308–13313 (2003).
Tsuji, A. et al. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J. 78, 3260–3274 (2000).
Prigodich, A. E. et al. Nano-flares for mRNA regulation and detection. ACS Nano 3, 2147–2152 (2009).
Lee, S. E., Liu, G. L., Kim, F. & Lee, L. P. Remote optical switch for localized and selective control of gene interference. Nano Lett. 9, 562–570 (2009).
Seferos, D. S., Giljohann, D. A., Hill, H. D., Prigodich, A. E. & Mirkin, C. A. Nano-flares: probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 129, 15477–15479 (2007).
Dubois, A., Canva, M., Brun, A., Chaput, F. & Boilot, J-P. Photostability of dyemolecules trapped in solid matrices. Appl. Opt. 35, 3193–3199 (1996).
Jun, Y-w. et al. Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level. Proc. Natl Acad. Sci. USA 106, 17735–17740 (2009).
Cognet, L. et al. Single metallic nanoparticle imaging for protein detection in cells. Proc. Natl Acad. Sci. USA 100, 11350–11355 (2003).
Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996).
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules' using DNA. Nature 382, 609–611 (1996).
Elghanian, R., Storhoff, J. J., Mucic, R. C., Letsinger, R. L. & Mirkin, C. A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277, 1078–1081 (1997).
Yguerabide, J. & Yguerabide, E. E. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications: II. Experimental characterization. Anal. Biochem. 262, 157–176 (1998).
Lee, K. & Irudayaraj, J. Periodic and dynamic 3-D gold nanoparticle–DNA network structures for surface-enhanced Raman spectroscopy-based quantification. J. Phys. Chem. C 113, 5980–5983 (2009).
Graham, D., Thompson, D. G., Smith, W. E. & Faulds, K. Control of enhanced Raman scattering using a DNA-based assembly process of dye-coded nanoparticles. Nature Nanotech. 3, 548–551 (2008).
Bao-An, D., Zheng-Ping, L. & Cheng-Hui, L. One-step homogeneous detection of DNA hybridization with gold nanoparticle probes by using a linear light-scattering technique. Angew. Chem. Int. Ed. 45, 8022–8025 (2006).
Storhoff, J. J., Lucas, A. D., Garimella, V., Bao, Y. P. & Muller, U. R. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nature Biotechnol. 22, 883–887 (2004).
Taton, T. A., Mirkin, C. A. & Letsinger, R. L. Scanometric DNA array detection with nanoparticle probes. Science 289, 1757–1760 (2000).
De La Rica, R. & Stevens, M. M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nature Nanotech. 7, 821–824 (2012).
Reinhard, B. r. M., Sheikholeslami, S., Mastroianni, A., Alivisatos, A. P. & Liphardt, J. Use of plasmon coupling to reveal the dynamics of DNA bending and cleavage by single EcoRV restriction enzymes. Proc. Natl Acad. Sci. USA 104, 2667–2672 (2007).
Sonnichsen, C., Reinhard, B. M., Liphardt, J. & Alivisatos, A. P. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nature Biotechnol. 23, 741–745 (2005).
Liu, G. L. et al. A nanoplasmonic molecular ruler for measuring nuclease activity and DNA footprinting. Nature Nanotech. 1, 47–52 (2006).
Raschke, G. et al. Biomolecular recognition based on single gold nanoparticle light scattering. Nano Lett. 3, 935–938 (2003).
Lee, K. & Irudayaraj, J. Correct spectral conversion between surface-enhanced Raman and plasmon resonance scattering from nanoparticle dimers for single-molecule detection. Small 9, 1106–1115 (2012).
Lee, K., Drachev, V. P. & Irudayaraj, J. DNA–gold nanoparticle reversible networks grown on cell surface marker sites: application in diagnostics. ACS Nano 5, 2109–2117 (2011).
Jain, P. K., Huang, W. & El-Sayed, M. A. On the universal scaling behavior of the distance decay of plasmon coupling in metal nanoparticle pairs: a plasmon ruler equation. Nano Lett. 7, 2080–2088 (2007).
Mie, G. Contributions to the optics of diffusing media. Ann. Physik 25, 377 (1908).
Debye, P. Light scattering in solutions. J. Appl. Phys. 15, 338–342 (1944).
Capecchi, M. R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 22, 479–488 (1980).
Feng, L., Huang, J. & Chen, J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 (2009).
Wang, B., Hurov, K., Hofmann, K. & Elledge, S. J. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 23, 729–739 (2009).
Shao, G. et al. MERIT40 controls BRCA1–Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
Orban, T. I. & Olah, E. Emerging roles of BRCA1 alternative splicing. Mol. Pathol. 56, 191–197 (2003).
Sun, L. & Irudayaraj, J. PCR-free quantification of multiple splice variants in a cancer gene by surface-enhanced Raman spectroscopy. J. Phys. Chem. B 113, 14021–14025 (2009).
Sun, L. & Irudayaraj, J. Quantitative surface-enhanced Raman for gene expression estimation. Biophys. J. 96, 4709–4716 (2009).
Sun, L., Yu, C. & Irudayaraj, J. Raman multiplexers for alternative gene splicing. Anal. Chem. 80, 3342–3349 (2008).
Seetin, M. G. & Mathews, D. H. in Bacterial Regulatory RNA 99–122 (Springer, 2012).
Rouskin, S., Zubradt, M., Washietl, S., Kellis, M. & Weissman, J. S. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2013).
Krude, T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp. Cell Res. 247, 148–159 (1999).
Grunwald, D. & Singer, R. H. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607 (2010).
Larson, D. R., Zenklusen, D., Wu, B., Chao, J. A. & Singer, R. H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332, 475–478 (2011).
Yu, C., Nakshatri, H. & Irudayaraj, J. Identity profiling of cell surface markers by multiplex gold nanorod probes. Nano Lett. 7, 2300–2306 (2007).
Acknowledgements
The authors thank Z. Machaty and K. Lee for the initial discussion on microinjection. The authors acknowledge funding to J.I. from the National Science Foundation (award no. 1249315), the Indiana Clinical Translational Sciences Institute (NIH-TR000006) and the Purdue Center for Cancer Research (NIH-NCI CCSG CA23168) and funding to K.L. and L.L. from Samsung (GRO 20122166) and Stanford NIH (award no. U54CA151459).
Author information
Authors and Affiliations
Contributions
K.L. and J.I. conceived the original idea for this study. K.L., L.L. and J.I. designed the experiments. K.L. built the hyperspectral imaging system and developed the data analysis algorithm. K.L. performed the nanoparticle dimer probe design/synthesis/characterization, microinjection and optical measurements. Y.C. performed the PCR, flow cytometry and other supporting biological validations. K.L., L.L. and J.I. analysed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 2141 kb)
Rights and permissions
About this article
Cite this article
Lee, K., Cui, Y., Lee, L. et al. Quantitative imaging of single mRNA splice variants in living cells. Nature Nanotech 9, 474–480 (2014). https://doi.org/10.1038/nnano.2014.73
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2014.73
This article is cited by
-
Phase intensity nanoscope (PINE) opens long-time investigation windows of living matter
Nature Communications (2023)
-
Tracking endocytosis and intracellular distribution of spherical nucleic acids with correlative single-cell imaging
Nature Protocols (2021)
-
High-spatial and colourimetric imaging of histone modifications in single senescent cells using plasmonic nanoprobes
Nature Communications (2021)
-
Plasmonic gold nanostructures for biosensing and bioimaging
Microchimica Acta (2021)
-
Nanotechnology in cancer diagnosis: progress, challenges and opportunities
Journal of Hematology & Oncology (2019)